Proteases
Proteases are the major pathway in the degradation of misfolded protein in protein synthesis and other proteolyzed protein, which are found in all eukaryotic cells, archaea, and some bacteria. Proteases, composed of plural Catalytic protein and regulatory protein, have multiple catalytic activities. It has three to four kinds of different peptidase activities, including the hydrolysis activity of tryptase, chymotrypsin-like enzyme and peptidylglutamyl-peptide. Depending on the sedimentation coefficient, it can be divided into two types of proteases: the 26S proteasome, of which the molecular weight is about 2000 kDa, dependent on ATP; and 20S proteasome, of which relative molecular mass of about 750 kDa, independent on ATP. The degradation of most protein by the proteasome requires ubiquitination.
26s proteasome is a multi-subunit protease of 2-MDa, which can degrade most of the cytoplasm, endoplasmic reticulum and nucleoprotein. The most understood target protein degradation mechanism is ubiquitin degradation via ubiquitin, an 8-kDa protein. 26s proteasome is composed of a core protein of 20s and the regulatory complex composition of 19s .The chaperone of 19s can unfold ubiquitin-labeled protein substrate and send it into the graphics core of 20s, where there are three pairs of proteolytic active sites, whose names are corresponding to the specific similarity to the cleavage specificity of chymotrypsin, trypsin and caspase. Studies on bud yeast mutants ultimately suggest the different role of these three pairs of sites. And the chymotrypsin-like active sites are most crucial to normal and regular growth. Proteasome inhibitors have been applied in the treatment of diseases such as cancer as well as widely used in laboratory studies. MG-341 is an FDA-approved medication for multiple myeloma and mantle cell lymphoma. It can also be used in laboratory studies to inhibit the activity of proteasome. Other protease inhibitors including disulfiram, epigallocatechin-3-gallate, salinosporamide A, carfilzomib, ONX0912, CEP-18770 and MLN9708 have also been developed or are in clinical trials for drugs and testing phase.
MG132, also known as Carbobenzoxy-L-leucyl-L-leucyl-L-leucinal, Z-LLL-CHO, is a peptide aldehyde, capable of inhibiting the activity of different types of proteases, including serine proteases and calpains. MG132 and other peptide aldehydes can effectively inhibit the protease activity of several peptidases in Proteasome as well as the activity of calpains.
- Structure:
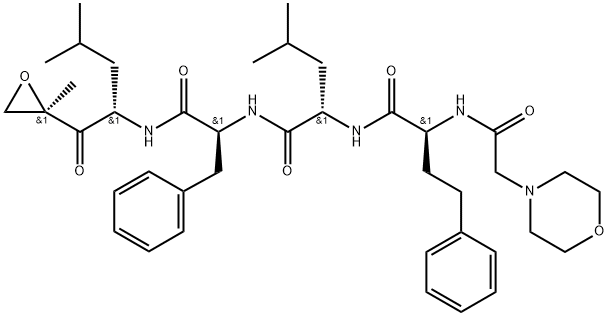
- Chemical Name:Carfilzomib
- CAS:868540-17-4
- MF:C40H57N5O7
- Structure:
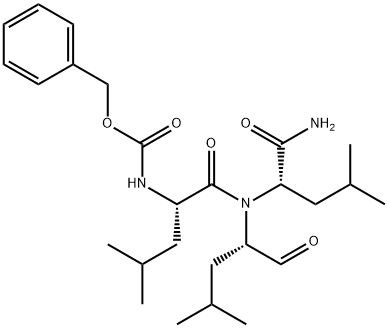
- Chemical Name:MG-132
- CAS:133407-82-6
- MF:C26H41N3O5
- Structure:
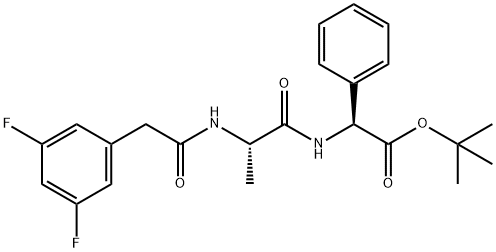
- Chemical Name:DAPT
- CAS:208255-80-5
- MF:C23H26F2N2O4
- Structure:
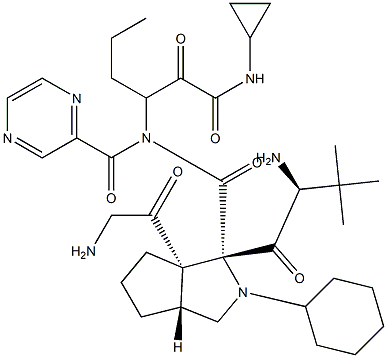
- Chemical Name:Telaprevir
- CAS:402957-28-2
- MF:C36H53N7O6
- Structure:
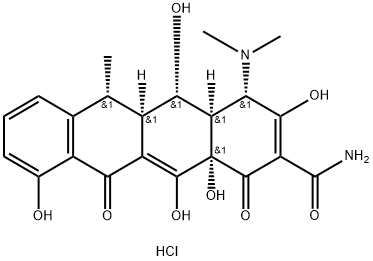
- Chemical Name:Doxycycline hydrochloride
- CAS:10592-13-9
- MF:C22H25ClN2O8
- Structure:
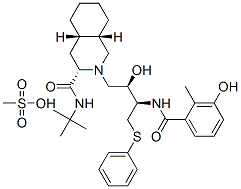
- Chemical Name:Nelfinavir mesylate
- CAS:159989-65-8
- MF:C33H49N3O7S2
- Structure:
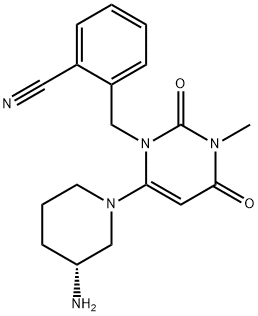
- Chemical Name:ALOGLIPTIN(ALOGLIPTINE, ALOGLIPTINA)
- CAS:850649-61-5
- MF:C18H21N5O2
- Structure:
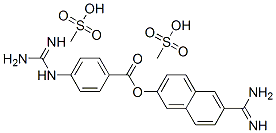
- Chemical Name:Nafamostat mesylate
- CAS:82956-11-4
- MF:C21H25N5O8S2
- Structure:
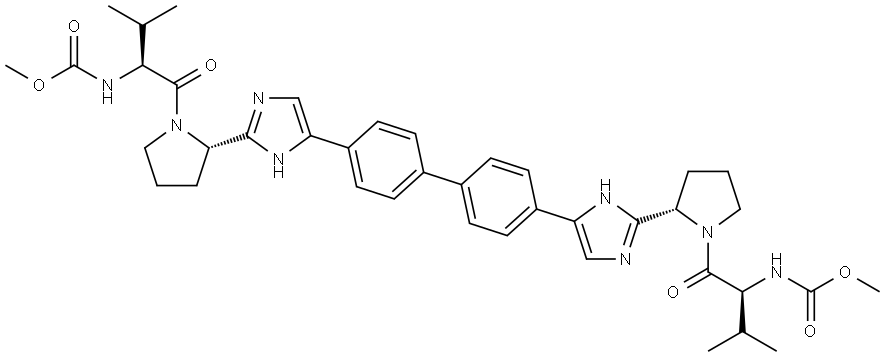
- Chemical Name:Daclatasvir
- CAS:1009119-64-5
- MF:C40H50N8O6
- Structure:
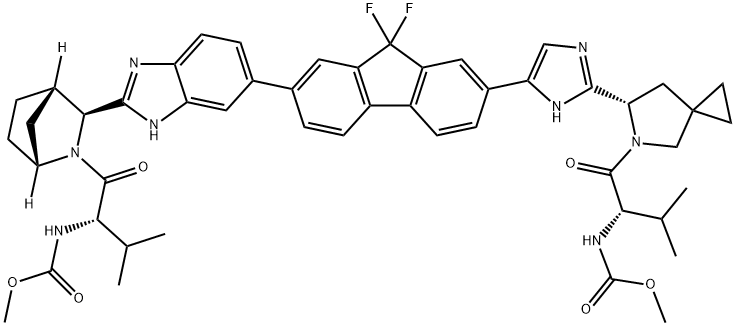
- Chemical Name:Ledipasvir
- CAS:1256388-51-8
- MF:C49H54F2N8O6
- Structure:
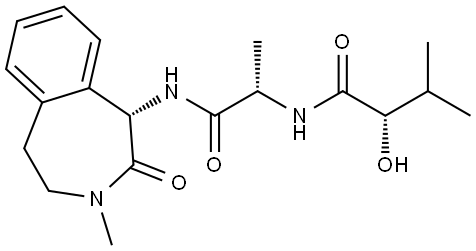
- Chemical Name:Semagacestat
- CAS:425386-60-3
- MF:C19H27N3O4
- Structure:
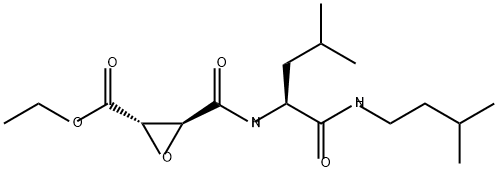
- Chemical Name:EST
- CAS:88321-09-9
- MF:C17H30N2O5
- Structure:
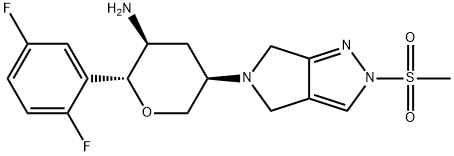
- Chemical Name:Omarigliptin (MK-3102)
- CAS:1226781-44-7
- MF:C17H20F2N4O3S
- Structure:
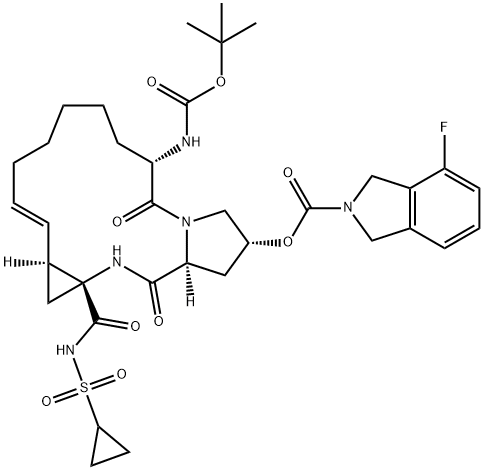
- Chemical Name:Danoprevir
- CAS:850876-88-9
- MF:C35H46FN5O9S
- Structure:
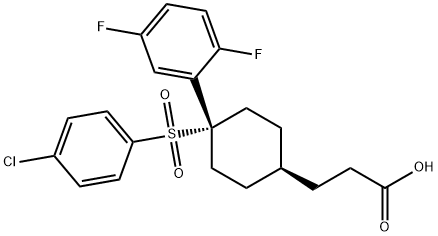
- Chemical Name:cis-4-[(4-Chlorophenyl)sulfonyl]-4-(2,5-difluorophenyl)cyclohexanepropanoic acid
- CAS:471905-41-6
- MF:C21H21ClF2O4S
- Structure:
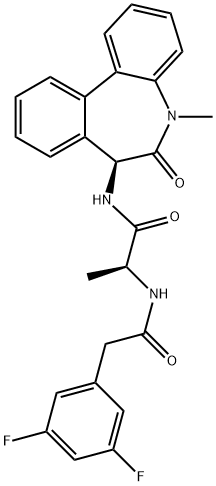
- Chemical Name:DIBENZAZEPINE
- CAS:209984-56-5
- MF:C26H23F2N3O3
- Structure:
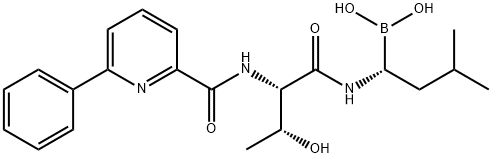
- Chemical Name:CEP-18770
- CAS:847499-27-8
- MF:C21H28BN3O5
- Structure:
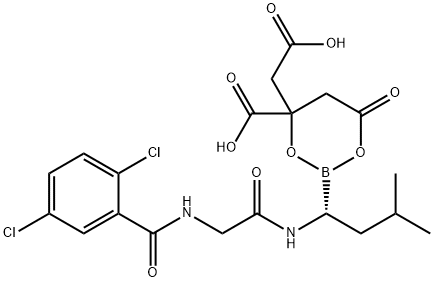
- Chemical Name:MLN9708
- CAS:1201902-80-8
- MF:C20H23BCl2N2O9
- Structure:
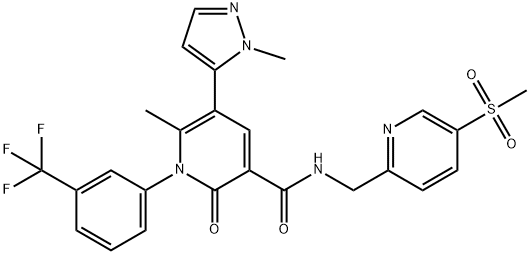
- Chemical Name:AZD 9668
- CAS:848141-11-7
- MF:C25H22F3N5O4S
- Structure:
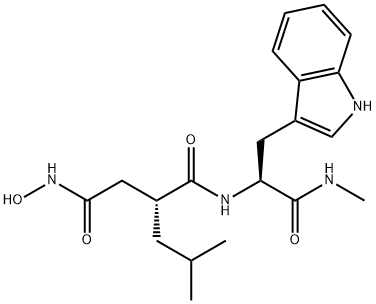
- Chemical Name:Ilomastat
- CAS:142880-36-2
- MF:C20H28N4O4
- Structure:
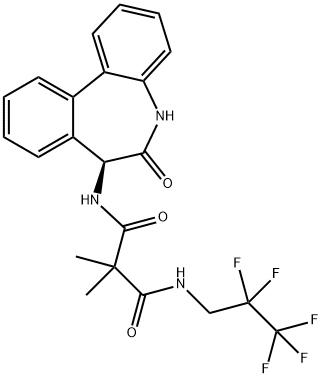
- Chemical Name:RO4929097
- CAS:847925-91-1
- MF:C22H20F5N3O3
- Structure:
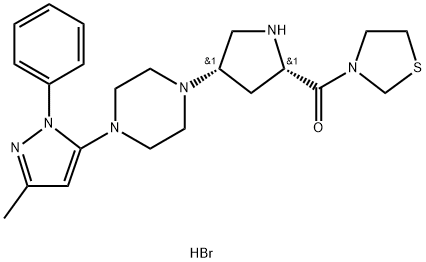
- Chemical Name:Teneligliptin Hydrobromide
- CAS:906093-29-6
- MF:C22H31BrN6OS
- Structure:
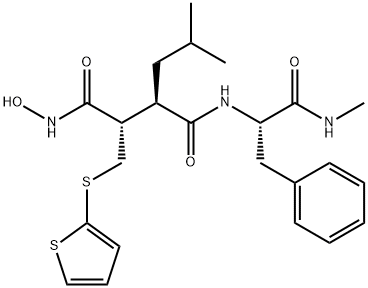
- Chemical Name:Batimastat
- CAS:130370-60-4
- MF:C23H31N3O4S2
- Structure:
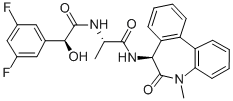
- Chemical Name:LY411575
- CAS:209984-57-6
- MF:C26H23F2N3O4
- Structure:
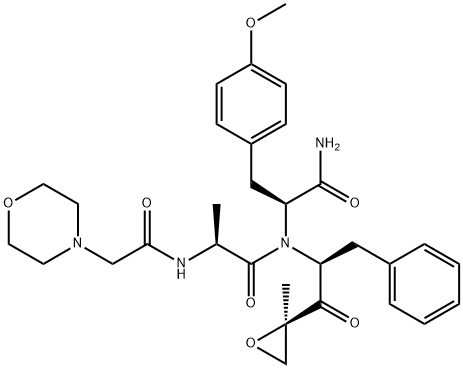
- Chemical Name:ONX-0914
- CAS:960374-59-8
- MF:C31H40N4O7
- Structure:
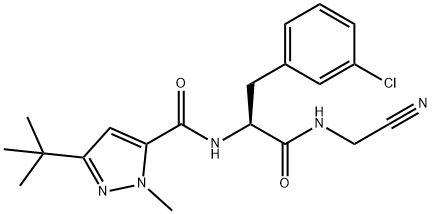
- Chemical Name:Cathepsin Inhibitor 1
- CAS:225120-65-0
- MF:C20H24ClN5O2
- Structure:

- Chemical Name:PD 151746
- CAS:179461-52-0
- MF:C11H8FNO2S
- Structure:
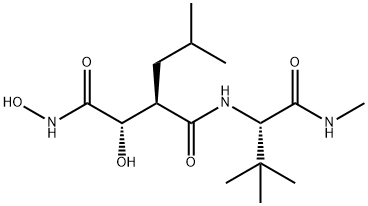
- Chemical Name:Marimastat
- CAS:154039-60-8
- MF:C15H29N3O5
- Structure:
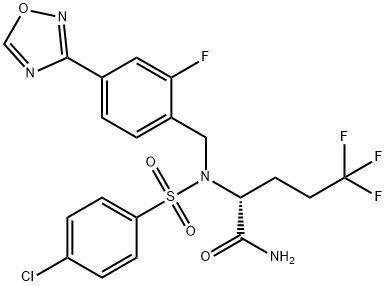
- Chemical Name:Avagacestat (BMS-708163)
- CAS:1146699-66-2
- MF:C20H17ClF4N4O4S
- Structure:
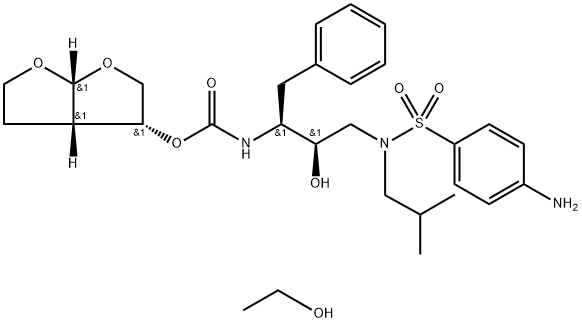
- Chemical Name:Darunavir ethanolate
- CAS:635728-49-3
- MF:C29H43N3O8S
- Structure:
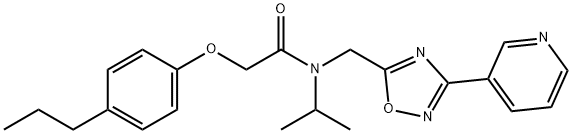
- Chemical Name:PI-1840
- CAS:1401223-22-0
- MF:C22H26N4O3
- Structure:
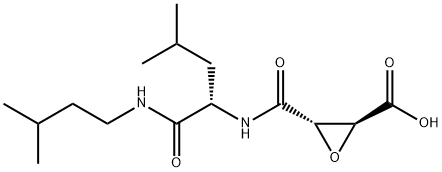
- Chemical Name:E-64C
- CAS:76684-89-4
- MF:C15H26N2O5
- Structure:
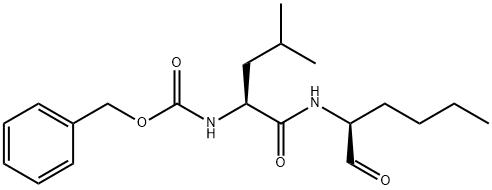
- Chemical Name:CALPEPTIN
- CAS:117591-20-5
- MF:C20H30N2O4
- Structure:
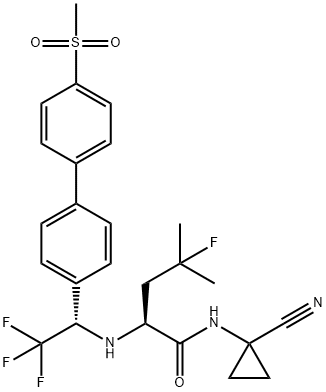
- Chemical Name:MK-0822
- CAS:603139-19-1
- MF:C25H27F4N3O3S
- Structure:
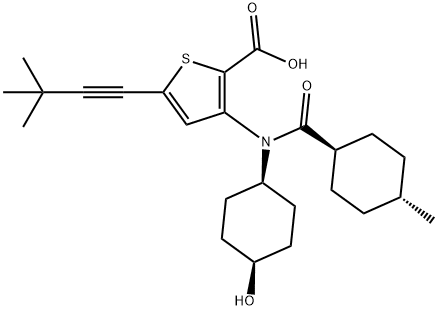
- Chemical Name:VX-222
- CAS:1026785-59-0
- MF:C25H35NO4S
- Structure:
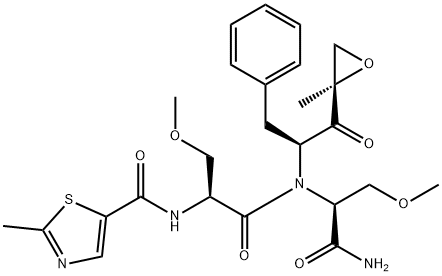
- Chemical Name:Oprozomib
- CAS:935888-69-0
- MF:C25H32N4O7S
- Structure:
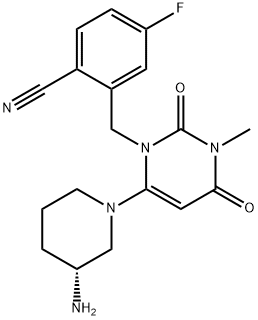
- Chemical Name:Trelagliptin
- CAS:865759-25-7
- MF:C18H20FN5O2
- Structure:
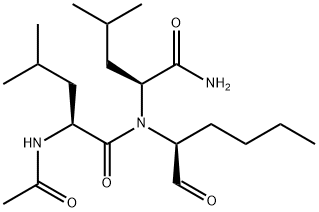
- Chemical Name:Calpain Inhibitor I
- CAS:110044-82-1
- MF:C20H37N3O4
- Structure:
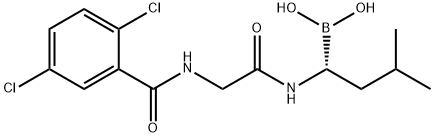
- Chemical Name:(R)-1-(2-(2,5-dichlorobenzamido)acetamido)-3-methylbutylboronic acid
- CAS:1072833-77-2
- MF:C14H19BCl2N2O4
- Structure:
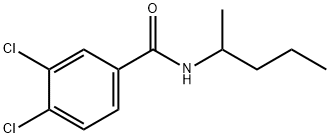
- Chemical Name:3,4-Dichloro-N-(1-Methylbutyl)benza-Mide
- CAS:7497-07-6
- MF:C12H15Cl2NO
- Structure:
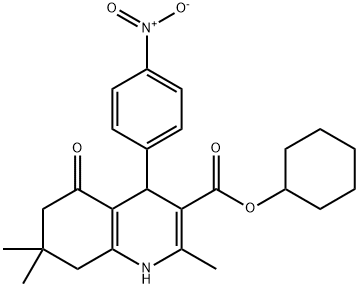
- Chemical Name:FLI-06
- CAS:313967-18-9
- MF:C25H30N2O5
- Structure:
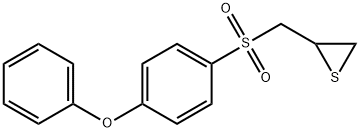
- Chemical Name:MMP-2/MMP-9 INHIBITOR IV
- CAS:292605-14-2
- MF:C15H14O3S2
- Structure:
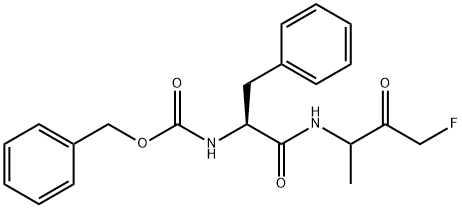
- Chemical Name:Z-PHE-DL-ALA-FLUOROMETHYLKETONE
- CAS:197855-65-5
- MF:C21H23FN2O4