PI3K / Akt / mTOR
The phosphatidylinositol 3-kinase (PI3Ks) family of proteins participates into the regulation of cell survival, growth, metabolism and glucose homeostasis and other cellular functions. The increased PI3K kinase activity is often associated with various kinds of cancers. As the name suggests, PI3Ks can phosphorylate the 3-position carbon atom of the inositol ring in the phosphatidylinositol PI (a kind of membrane phospholipids). PI has a very small fraction in the cell membrane with its content being less than phosphatidylcholine phosphatidylethanolamine and phosphatidylserine. However, it is abundant in the brain cells with its content reaching up to 10% of the total phospholipids.
PI3K activation
PI3K can be divided into three categories with different structures and functions. The most extensively studied PI3K is the class I PI3K. This kind of PI3K is heterodimer consisting of a regulatory subunit and a catalytic subunit. Regulatory subunits contain SH2 and SH3 domain and can interact with the related target protein containing the corresponding binding site. The subunit is usually called p85, referred to the first found subtype (isotype). However, the currently known six regulatory subunits have the size ranging from 50 to 110kDa. There are four kinds of catalytic subunits, namely p110α, β, δ, γ with δ being limited to only white blood cells but the rest being widely distributed in various cells.
The activation of PI3K is largely relying on the substrate coming near to its part in the inner plasma membrane. There are various kinds of factors and signaling complexes, including fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF), human growth factor (of HGF), vascular ARNT I (Ang1) and insulin that can initiate the PI3K activation process.
These factors can activate the receptor tyrosine kinase (RTK), resulting in auto-phosphorylation. The phosphorylated residue in the receptor has provided a docking site for the heterologous dimerized PI3Kp85 subunits. However, in some cases, receptor phosphorylation is mediated by recruiting an adapter protein.
For example, when insulin activates its receptor, it must recruit a protein insulin receptor substrate (IRS) to facilitate binding of PI3K. Similarly, when the whole protein integrin (non-RTK) is activated, the focal adhesion kinase (FAK) can be used as the adapter protein, docking the PI3K through its p85 subunit. However, in the above case, the SH2 and SH3 domains of the p85 subunit are both connected with linker protein in a phosphorylation site. After PI3K recruits it to the activated receptors, it initiates the phosphorylation of multiple PI intermediates. PI3K, which is especially related with cancer, converts the PIP2 to PIP3.
The role of AKT
The activated AKT, through phosphorylation of various kinds of downstream enzymes, kinases and transcription factors, further regulates the cellular function. For example, AKT can stimulate the glucose metabolism: AKT can activate the AS160 (AKT substrate, 160 kDa), and further promote the absorption of GLUT4 transposition and the absorption of muscle cells on glucose. AKT can do phosphorylation of GSK3β to inhibit its activity, thereby promoting the glucose metabolism and regulating of cell cycle. AKT can phosphorylate the TSC 1/2 (tuberous sclerosis complex), being able to prevent its negative regulation on small G protein Rheb (Ras homology enriched in brain), thus causing Rheb enrichment and the activation of rapamycin-sensitive mTOR complexes (mTORC1). These effects can activate the protein translation, and enhance cell growth.
mTOR (mammalian target protein of rapamycin) kinase is a new member of the protein kinase family, this kind of protein kinase also belongs to phosphatidylinositol kinase-related kinase (PIKK). mTOR is found during the process of studying immunosuppressant rapamycin process. During the study, scientists found that immunosuppressant FK506 which is structurally similar with rapamycin can bind to the same target protein FKBP12 (FK506 binding protein) to exert its immunity inhibition. However, it has different mechanism of immunosuppression from FK506. The complex formed between rapamycin and FKBP12 can’t bind to calmodulin. Moreover, rapamycin can’t inhibit T cell activation or directly reduce the synthesis of early cytokine. It blocks the receptor signaling pathways through different cytokines, blocking the transition process of T lymphocytes and other cells from G1 to S phase. FK506, instead, block the proliferation of T lymphocytes from the G0 to G1 phase. Owing to the important position of mTOR in cell proliferation, differentiation, metastasis and survival, mTOR has become a new target in cancer therapy.
- Structure:
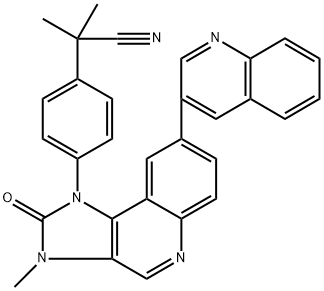
- Chemical Name:NVP-BEZ 235
- CAS:915019-65-7
- MF:C30H23N5O
- Structure:
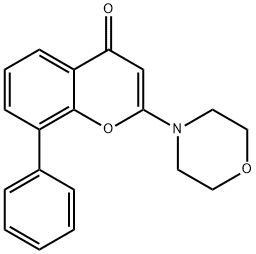
- Chemical Name:LY 294002 Hydrochloride
- CAS:154447-36-6
- MF:C19H17NO3
- Structure:
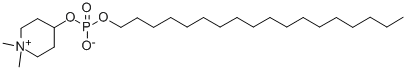
- Chemical Name:Perifosine (KRX-0401)
- CAS:157716-52-4
- MF:C25H52NO4P
- Structure:
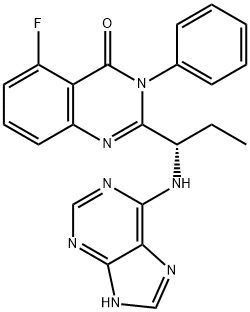
- Chemical Name:CAL-101
- CAS:870281-82-6
- MF:C22H18FN7O
- Structure:
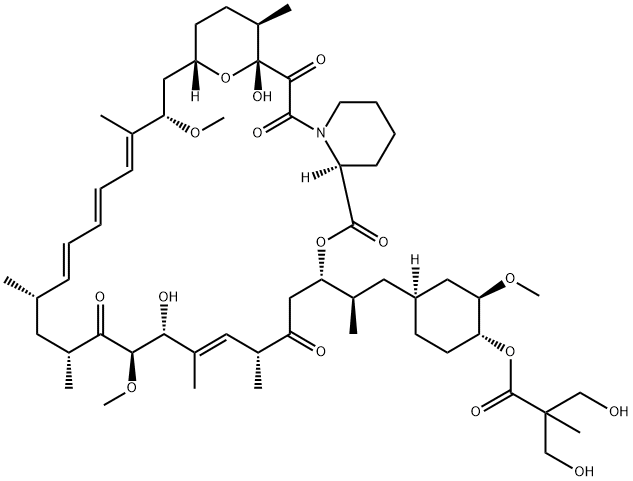
- Chemical Name:Temsirolimus
- CAS:162635-04-3
- MF:C56H87NO16
- Structure:
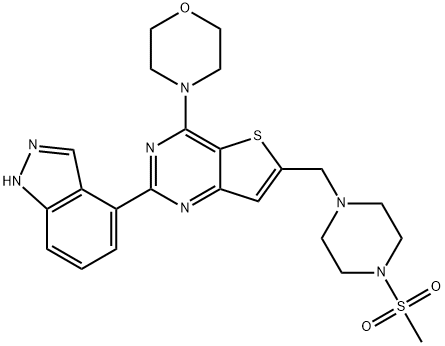
- Chemical Name:Pictilisib
- CAS:957054-30-7
- MF:C23H27N7O3S2
- Structure:
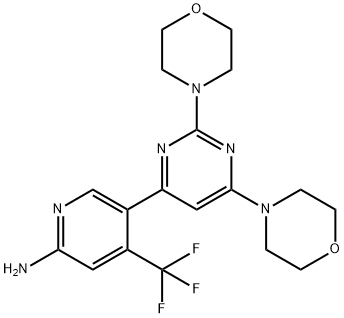
- Chemical Name:BKM120 (NVP-BKM120, Buparlisib)
- CAS:944396-07-0
- MF:C18H21F3N6O2
- Structure:
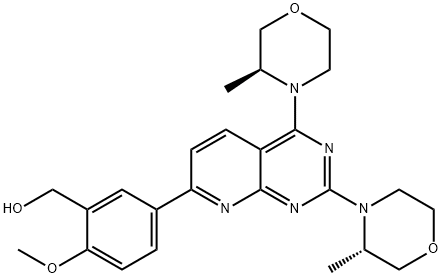
- Chemical Name:AZD-8055
- CAS:1009298-09-2
- MF:C25H31N5O4
- Structure:
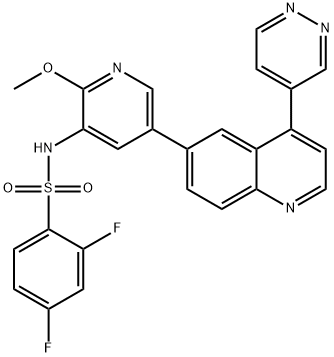
- Chemical Name:GSK-2126458
- CAS:1086062-66-9
- MF:C25H17F2N5O3S
- Structure:
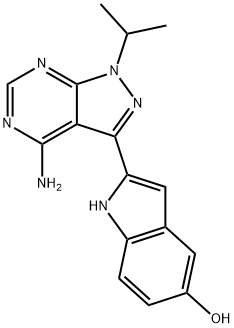
- Chemical Name:PP242
- CAS:1092351-67-1
- MF:C16H16N6O
- Structure:
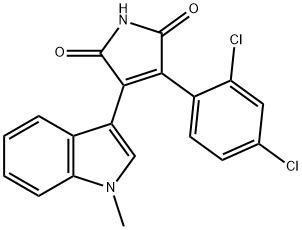
- Chemical Name:SB216763
- CAS:280744-09-4
- MF:C19H12Cl2N2O2
- Structure:
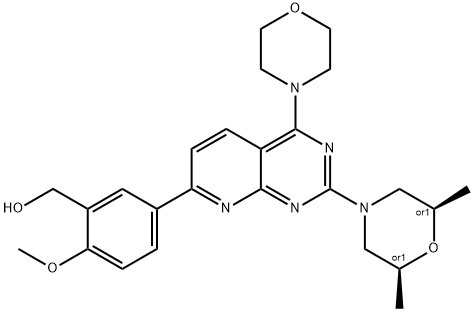
- Chemical Name:KU-0063794
- CAS:938440-64-3
- MF:C25H31N5O4
- Structure:
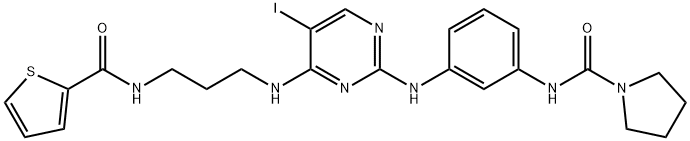
- Chemical Name:N-[3-[[5-Iodo-4-[[3-[(2-thienylcarbonyl)amino]propyl]amino]-2-pyrimidinyl]amino]phenyl]-1-pyrrolidinecarboxamide
- CAS:702675-74-9
- MF:C23H26IN7O2S
- Structure:
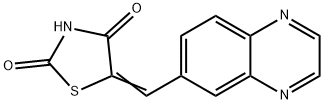
- Chemical Name:5-(6-Quinoxalinylmethylene)-2,4-thiazolidinedione
- CAS:648450-29-7
- MF:C12H7N3O2S
- Structure:
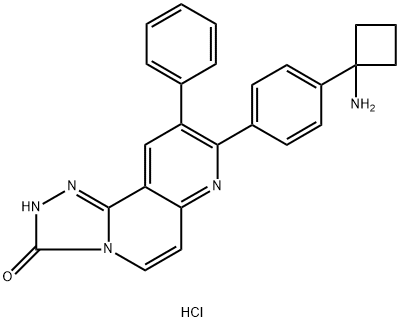
- Chemical Name:MK-2206 2HCl
- CAS:1032350-13-2
- MF:C25H22ClN5O
- Structure:
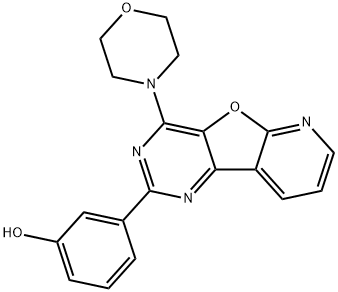
- Chemical Name:PI-103
- CAS:371935-74-9
- MF:C19H16N4O3
- Structure:
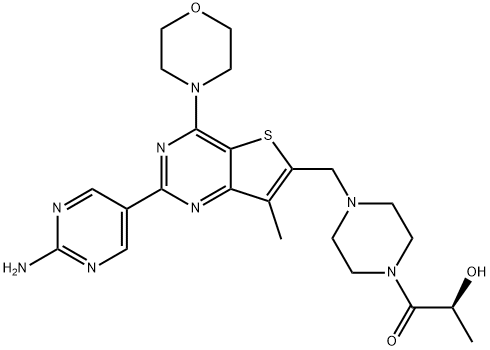
- Chemical Name:(S)-1-[4-[[2-(2-Aminopyrimidin-5-yl)-7-methyl-4-(morpholin-4-yl)thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-2-hydroxypropan-1-one
- CAS:1032754-93-0
- MF:C23H30N8O3S
- Structure:
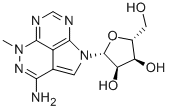
- Chemical Name:API-2
- CAS:35943-35-2
- MF:C13H16N6O4
- Structure:
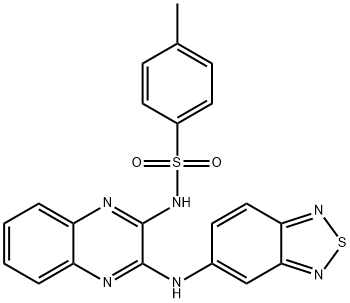
- Chemical Name:XL147
- CAS:956958-53-5
- MF:C21H16N6O2S2
- Structure:
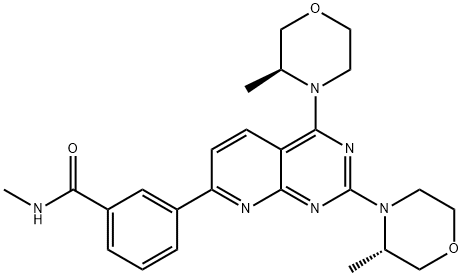
- Chemical Name:3-[2,4-Bis((3S)-3-methylmorpholin-4-yl)pyrido[5,6-e]pyrimidin-7-yl]-N-methylbenzamide
- CAS:1009298-59-2
- MF:C25H30N6O3
- Structure:
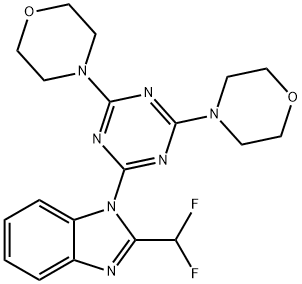
- Chemical Name:ZSTK474
- CAS:475110-96-4
- MF:C19H21F2N7O2
- Structure:
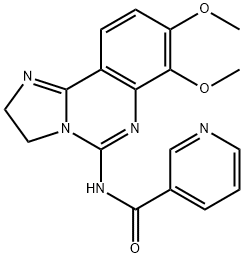
- Chemical Name:N-(2,3-Dihydro-7,8-dimethoxyimidazo[1,2-c]quinazolin-5-yl)-3-pyridinecarboxamide
- CAS:677338-12-4
- MF:C18H17N5O3
- Structure:
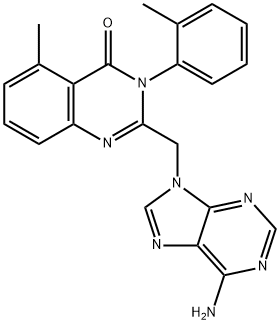
- Chemical Name:2-[(6-Amino-9H-purin-9-yl)methyl]-5-methyl-3-(2-methylphenyl)-4(3H)-quinazolinone
- CAS:371242-69-2
- MF:C22H19N7O
- Structure:
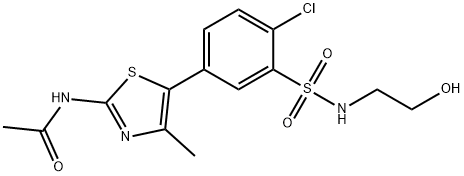
- Chemical Name:PIK-93
- CAS:593960-11-3
- MF:C14H16ClN3O4S2
- Structure:
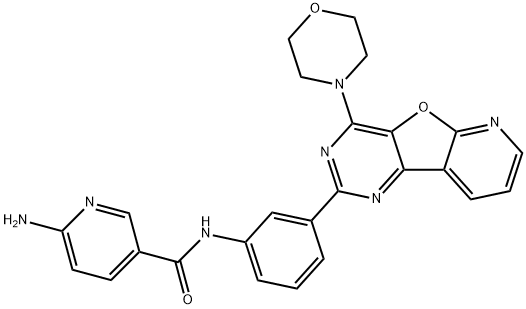
- Chemical Name:6-Amino-N-[3-[4-(4-morpholinyl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenyl]-3-pyridinecarboxamide
- CAS:371942-69-7
- MF:C25H21N7O3
- Structure:
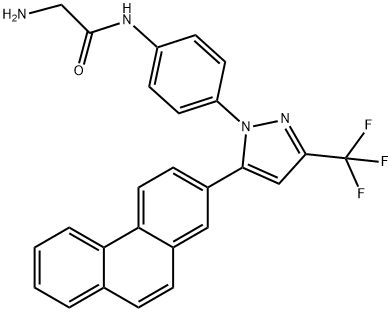
- Chemical Name:OSU-03012
- CAS:742112-33-0
- MF:C26H19F3N4O
- Structure:
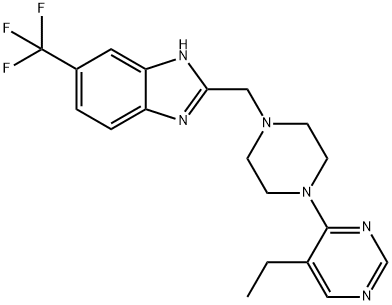
- Chemical Name:PF-4708671
- CAS:1255517-76-0
- MF:C19H21F3N6
- Structure:
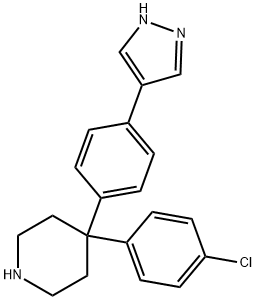
- Chemical Name:AT7867
- CAS:857531-00-1
- MF:C20H20ClN3
- Structure:
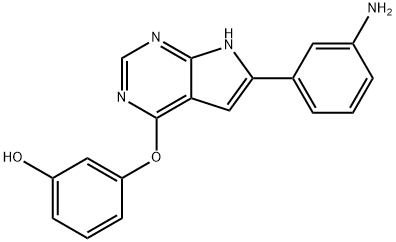
- Chemical Name:GSK-3BETA INHIBITOR XII
- CAS:601514-19-6
- MF:C18H14N4O2
- Structure:
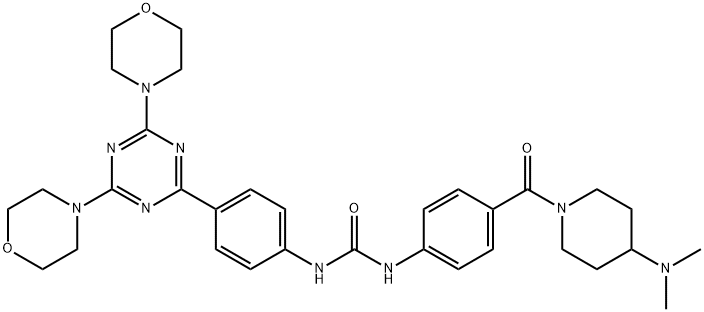
- Chemical Name:PF-05212384
- CAS:1197160-78-3
- MF:C32H41N9O4
- Structure:
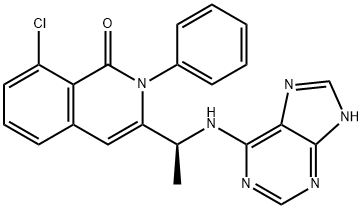
- Chemical Name:IPI 145
- CAS:1201438-56-3
- MF:C22H17ClN6O
- Structure:
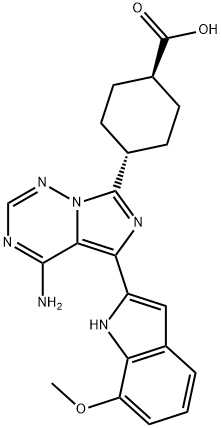
- Chemical Name:OSI-027
- CAS:936890-98-1
- MF:C21H22N6O3
- Structure:
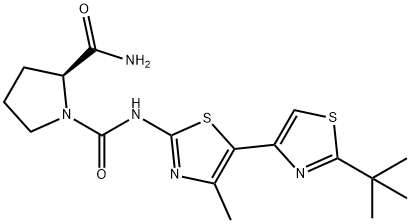
- Chemical Name:A66
- CAS:1166227-08-2
- MF:C17H23N5O2S2
- Structure:
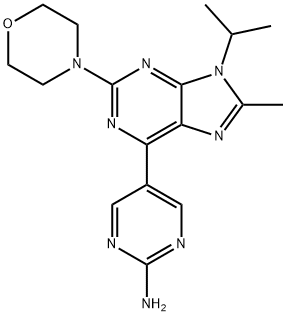
- Chemical Name:VS-5584
- CAS:1246560-33-7
- MF:C17H22N8O
- Structure:
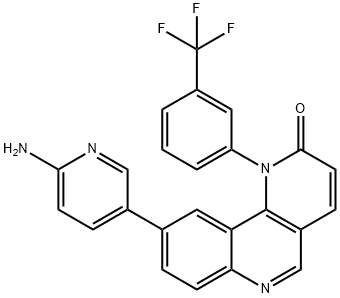
- Chemical Name:9-(6-Amino-3-pyridinyl)-1-[3-(trifluoromethyl)phenyl]benzo[h]-1,6-naphthyridin-2(1H)-one
- CAS:1223001-51-1
- MF:C24H15F3N4O
- Structure:
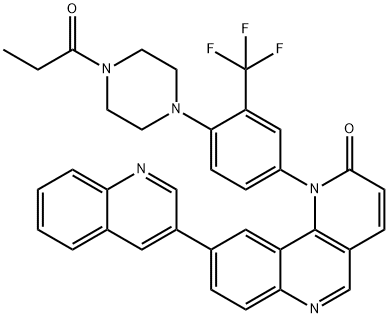
- Chemical Name:Torin 1
- CAS:1222998-36-8
- MF:C35H28F3N5O2
- Structure:
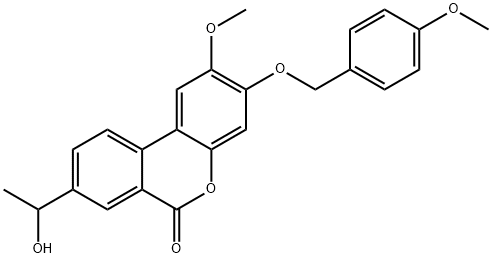
- Chemical Name:PaloMid 529 (P529)
- CAS:914913-88-5
- MF:C24H22O6
- Structure:
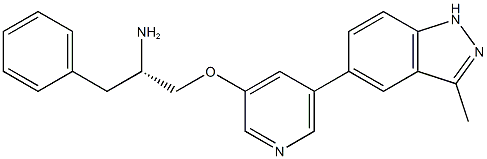
- Chemical Name:A-674563
- CAS:552325-73-2
- MF:C22H22N4O
- Structure:
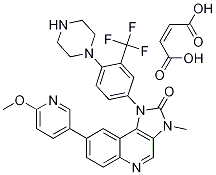
- Chemical Name:NVP-BGT226
- CAS:1245537-68-1
- MF:C28H25F3N6O2.C4H4O4
- Structure:
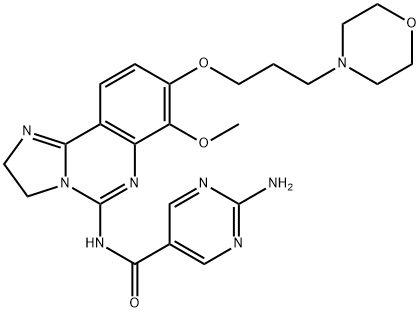
- Chemical Name:BAY 80-6946 (Copanlisib)
- CAS:1032568-63-0
- MF:C23H28N8O4
- Structure:
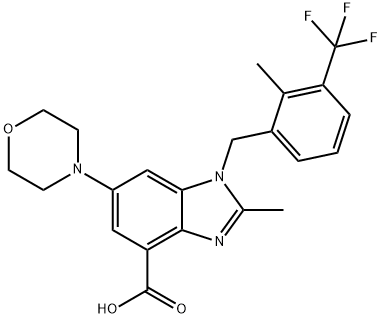
- Chemical Name:GSK2636771
- CAS:1372540-25-4
- MF:C22H22F3N3O3
- Structure:
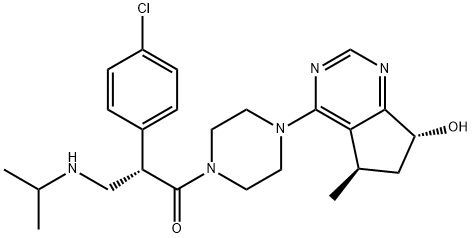
- Chemical Name:GDC-0068
- CAS:1001264-89-6
- MF:C24H32ClN5O2
- Structure:
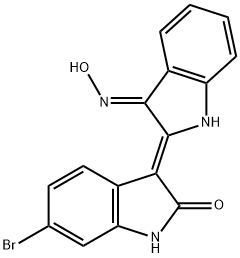
- Chemical Name:BIO
- CAS:667463-62-9
- MF:C16H10BrN3O2
- Structure:
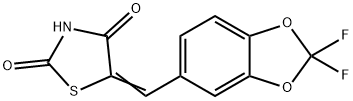
- Chemical Name:AS 604850
- CAS:648449-76-7
- MF:C11H5F2NO4S
- Structure:
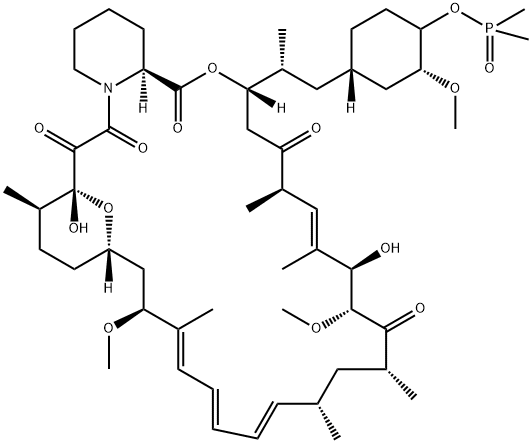
- Chemical Name:42-(Dimethylphosphinate)rapamycin
- CAS:572924-54-0
- MF:C53H84NO14P
- Structure:
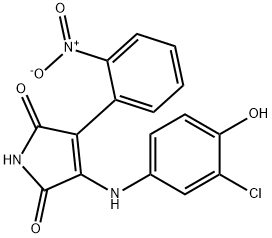
- Chemical Name:SB 415286
- CAS:264218-23-7
- MF:C16H10ClN3O5
- Structure:
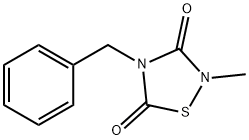
- Chemical Name:4-BENZYL-2-METHYL-1,2,4-THIADIAZOLIDINE-3,5-DIONE
- CAS:327036-89-5
- MF:C10H10N2O2S
- Structure:
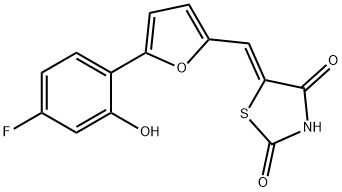
- Chemical Name:5-[[5-(4-Fluoro-2-hydroxyphenyl)-2-furanyl]methylene]-2,4-thiazolidinedione
- CAS:900515-16-4
- MF:C14H8FNO4S
- Structure:
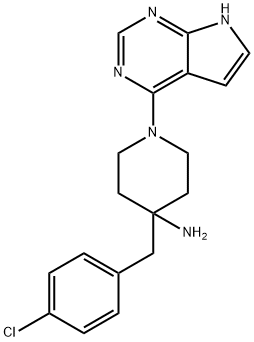
- Chemical Name:CCT128930
- CAS:885499-61-6
- MF:C18H20ClN5
- Structure:
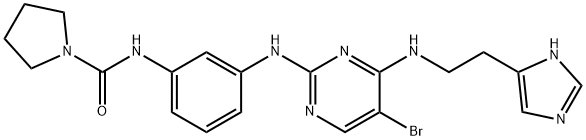
- Chemical Name:BX-912
- CAS:702674-56-4
- MF:C20H23BrN8O
- Structure:
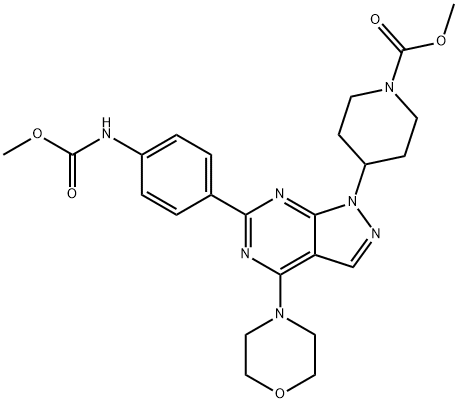
- Chemical Name:WYE-354
- CAS:1062169-56-5
- MF:C24H29N7O5
- Structure:
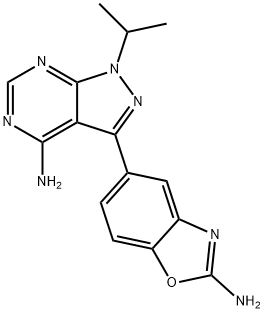
- Chemical Name:5-(4-aMino-1-isopropyl-1H-pyrazolo[3,4-d]pyriMidin-3-yl)benzo[d]oxazol-2-aMine
- CAS:1224844-38-5
- MF:C15H15N7O
- Structure:
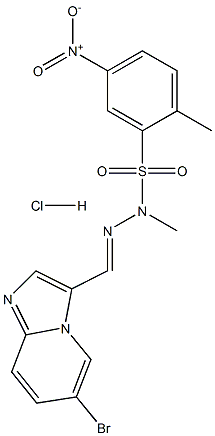
- Chemical Name:PIK-75 Hydrochloride
- CAS:372196-77-5
- MF:C16H15BrClN5O4S
- Structure:
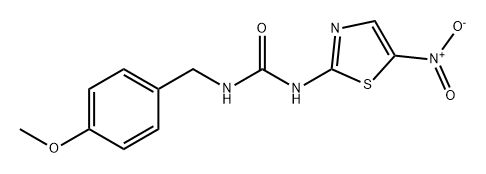
- Chemical Name:AR-A014418
- CAS:487021-52-3
- MF:C12H12N4O4S
- Structure:
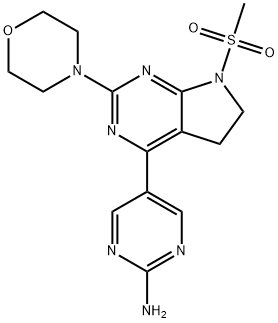
- Chemical Name:CH5132799
- CAS:1007207-67-1
- MF:C15H19N7O3S
- Structure:
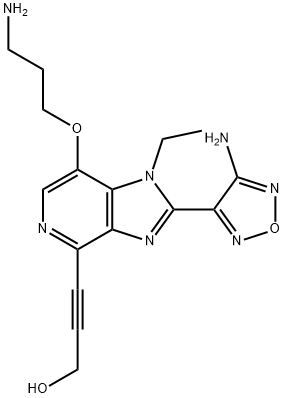
- Chemical Name:AKT Kinase Inhibitor
- CAS:842148-40-7
- MF:C16H19N7O3
- Structure:
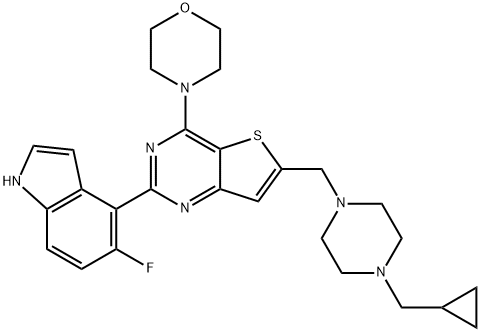
- Chemical Name:PI-3065
- CAS:955977-50-1
- MF:C27H31FN6OS
- Structure:
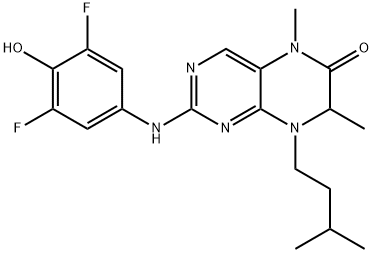
- Chemical Name:BI-D1870
- CAS:501437-28-1
- MF:C19H23F2N5O2
- Structure:
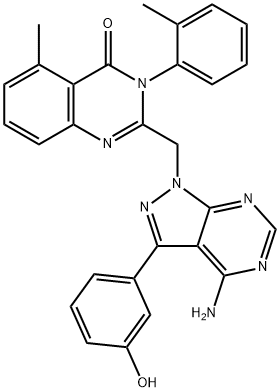
- Chemical Name:PIK-294
- CAS:900185-02-6
- MF:C28H23N7O2
- Structure:
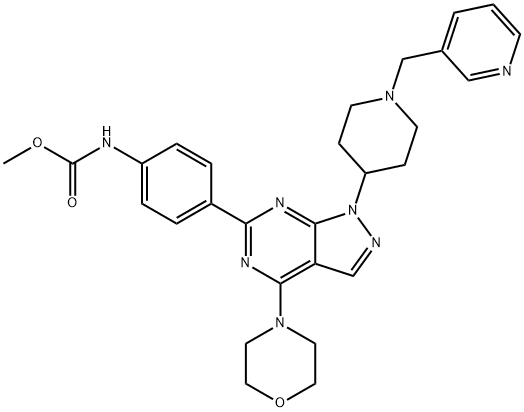
- Chemical Name:WYE-687
- CAS:1062161-90-3
- MF:C28H32N8O3
- Structure:
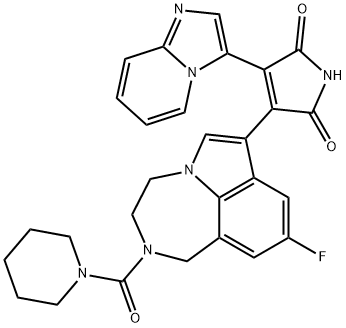
- Chemical Name:LY2090314
- CAS:603288-22-8
- MF:C28H25FN6O3
- Structure:
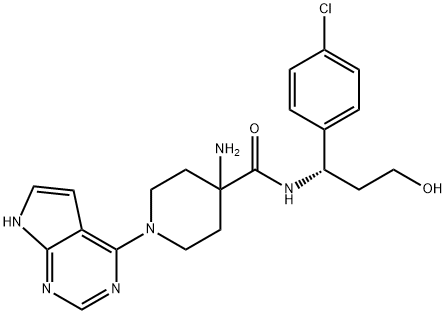
- Chemical Name:Capivasertib
- CAS:1143532-39-1
- MF:C21H25ClN6O2
- Structure:
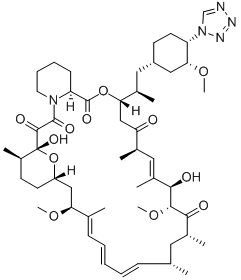
- Chemical Name:ZOTAROLIMUS
- CAS:221877-54-9
- MF:C52H79N5O12
- Structure:
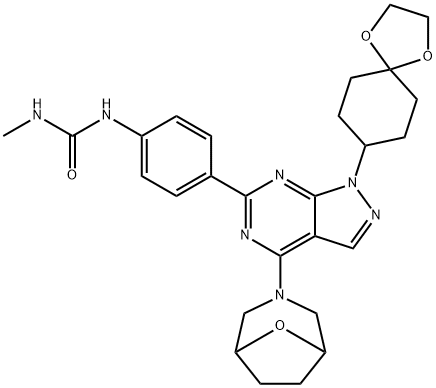
- Chemical Name:N-[4-[1-(1,4-Dioxaspiro[4.5]dec-8-yl)-4-(8-oxa-3-azabicyclo[3.2.1]oct-3-yl)-1H-pyrazolo[3,4-d]pyrimidin-6-yl]phenyl]-N'-methylurea
- CAS:1144068-46-1
- MF:C27H33N7O4
- Structure:
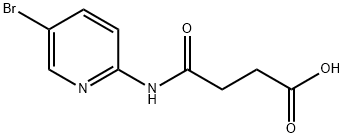
- Chemical Name:Bikinin
- CAS:188011-69-0
- MF:C9H9BrN2O3
- Structure:
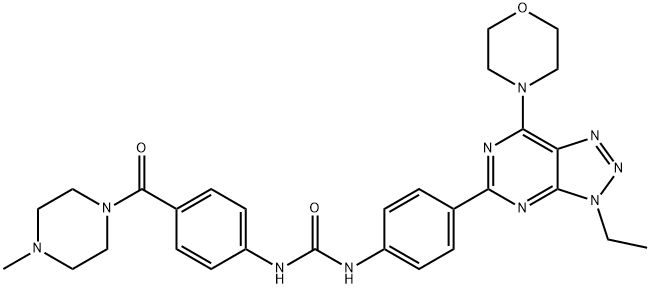
- Chemical Name:PKI-402
- CAS:1173204-81-3
- MF:C29H34N10O3
- Structure:
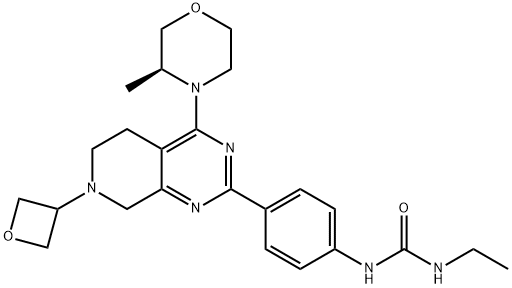
- Chemical Name:N-Ethyl-N'-[4-[5,6,7,8-tetrahydro-4-[(3S)-3-methyl-4-morpholinyl]-7-(3-oxetanyl)pyrido[3,4-d]pyrimidin-2-yl]phenyl]urea
- CAS:1207360-89-1
- MF:C24H32N6O3
- Structure:
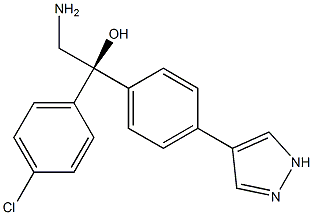
- Chemical Name:AT-13148
- CAS:1056901-62-2
- MF:C17H16ClN3O
- Structure:
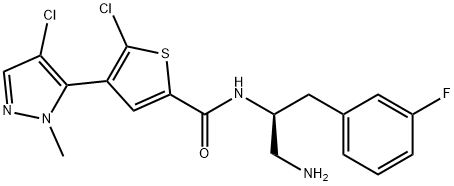
- Chemical Name:GSK-2110183C
- CAS:1047644-62-1
- MF:C18H17Cl2FN4OS
- Structure:
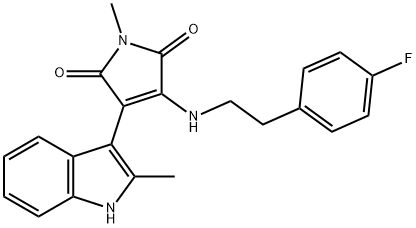
- Chemical Name:IM-12
- CAS:1129669-05-1
- MF:C22H20FN3O2
- Structure:
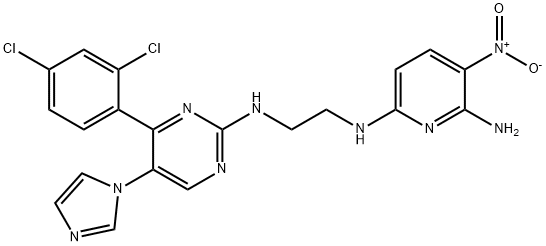
- Chemical Name:2,6-PYRIDINEDIAMINE, N6-[2-[[4-(2,4-DICHLOROPHENYL)-5-(1H-IMIDAZOL-1-YL)-2-PYRIMIDINYL]AMINO]ETHYL]-3-NITRO-
- CAS:252935-94-7
- MF:C20H17Cl2N9O2
- Structure:
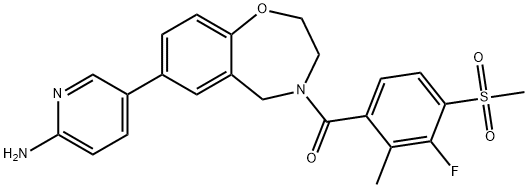
- Chemical Name:XL-388
- CAS:1251156-08-7
- MF:C23H22FN3O4S
- Structure:
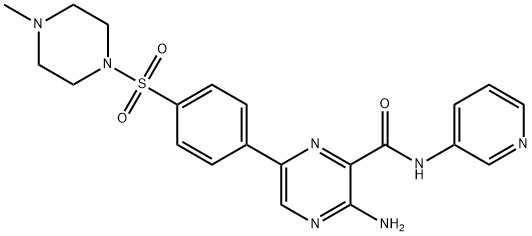
- Chemical Name:AZD-2858
- CAS:486424-20-8
- MF:C21H23N7O3S
- Structure:
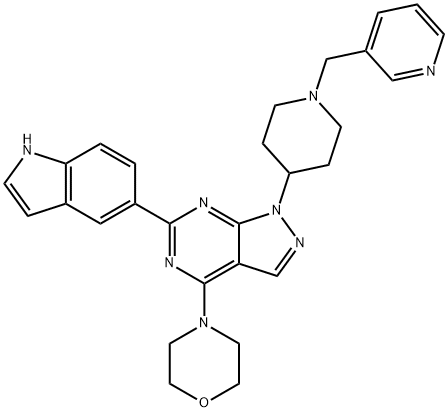
- Chemical Name:WAY-600
- CAS:1062159-35-6
- MF:C28H30N8O
- Structure:
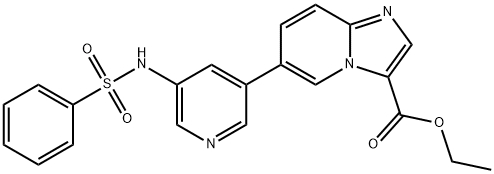
- Chemical Name:HS-173
- CAS:1276110-06-5
- MF:C21H18N4O4S
- Structure:
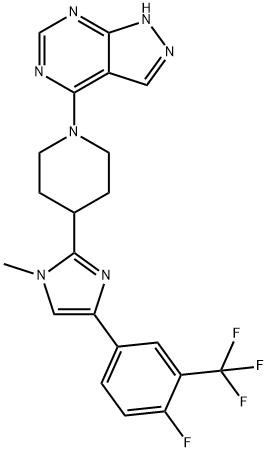
- Chemical Name:LY2584702
- CAS:1082949-67-4
- MF:C21H19F4N7
- Structure:
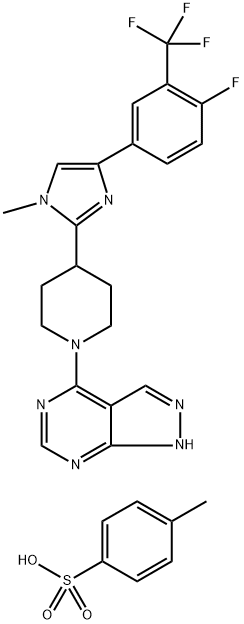
- Chemical Name:LY-2584702 (tosylate salt)
- CAS:1082949-68-5
- MF:C28H27F4N7O3S
- Structure:
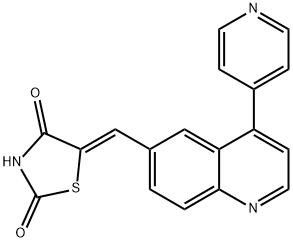
- Chemical Name:5-[[4-(4-Pyridinyl)-6-quinolinyl]methylene]-2,4-thiazolidenedione
- CAS:958852-01-2
- MF:C18H11N3O2S
- Structure:
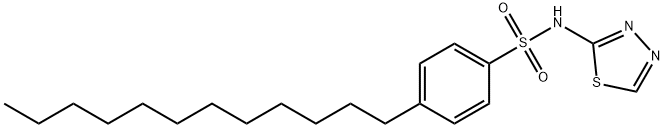
- Chemical Name:PHT-427
- CAS:1191951-57-1
- MF:C20H31N3O2S2
- Structure:
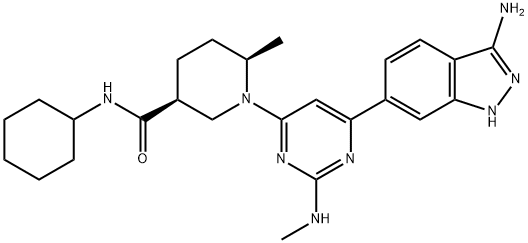
- Chemical Name:GSK 2334470
- CAS:1227911-45-6
- MF:C25H34N8O
- Structure:
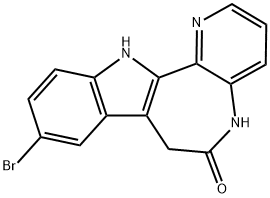
- Chemical Name:1-AZAKENPAULLONE
- CAS:676596-65-9
- MF:C15H10BrN3O
- Structure:
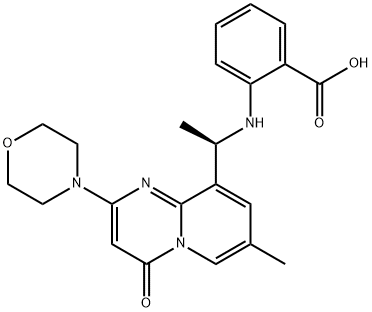
- Chemical Name:2-[[(1R)-1-[7-methyl-2-(4-morpholinyl)-4-oxo-4h-pyrido[1,2-a]pyrimidin-9-yl]ethyl]amino]benzoic acid
- CAS:1173900-33-8
- MF:C22H24N4O4
- Structure:
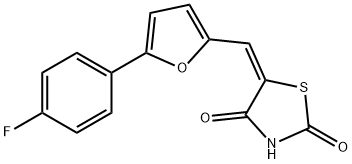
- Chemical Name:CAY10505
- CAS:1218777-13-9
- MF:C14H8FNO3S
- Structure:
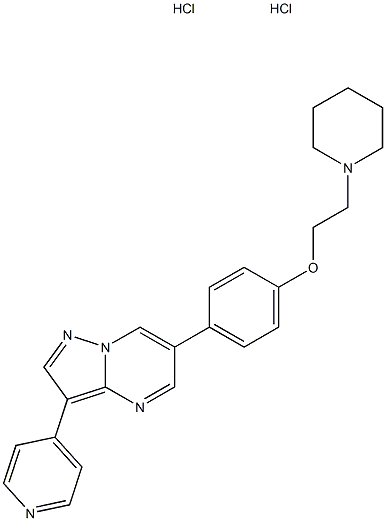
- Chemical Name:BML-275
- CAS:1219168-18-9
- MF:C24H27Cl2N5O
- Structure:
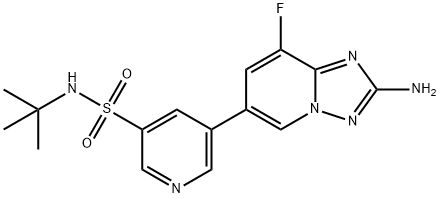
- Chemical Name:CZC24832
- CAS:1159824-67-5
- MF:C15H17FN6O2S
- Structure:
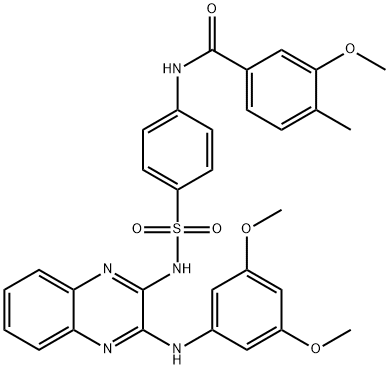
- Chemical Name:XL765
- CAS:1349796-36-6
- MF:C31H29N5O6S
- Structure:
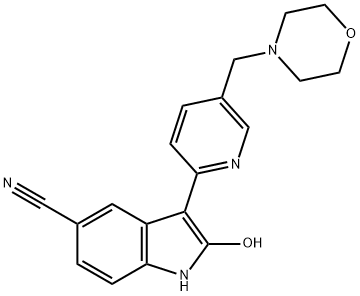
- Chemical Name:AZD 1080
- CAS:612487-72-6
- MF:C19H18N4O2
- Structure:
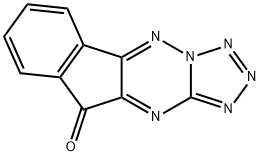
- Chemical Name:KP372-1
- CAS:329710-24-9
- MF:C10H4N6O
- Structure:
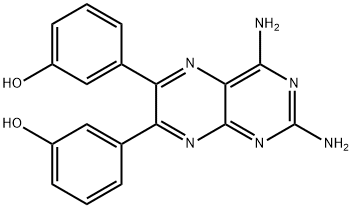
- Chemical Name:3,3'-(2,4-Diamino-6,7-pteridinediyl)bisphenol
- CAS:677297-51-7
- MF:C18H14N6O2
- Structure:
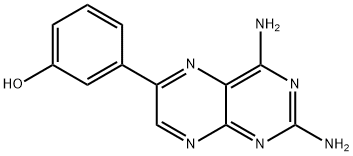
- Chemical Name:PIK-293
- CAS:900185-01-5
- MF:C22H19N7O
- Structure:
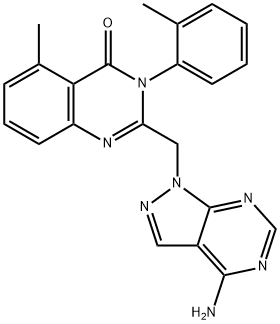
- Chemical Name:TG 100713
- CAS:925705-73-3
- MF:C12H10N6O
- Structure:
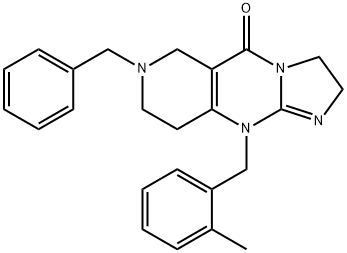
- Chemical Name:7-benzyl-10-(2-Methylbenzyl)-2,6,7,8,9,10-hexahydroiMidazo[1,2-a]pyrido[4,3-d]pyriMidin-5(3H)-one
- CAS:41276-02-2
- MF:C24H26N4O
- Structure:
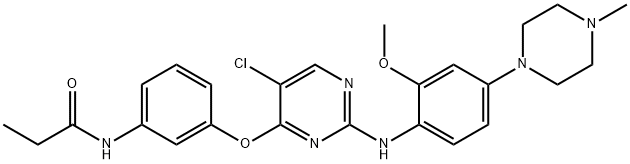
- Chemical Name:WZ4003
- CAS:1214265-58-3
- MF:C25H29ClN6O3
- Structure:
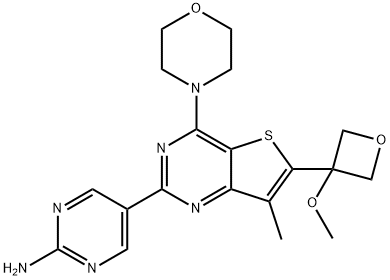
- Chemical Name:5-(6-(3-Methoxyoxetan-3-yl)-4-Morpholinothieno[3,2-d]pyriMidin-2-yl)pyriMidin-2-aMine
- CAS:1394076-92-6
- MF:C19H22N6O3S
- Structure:
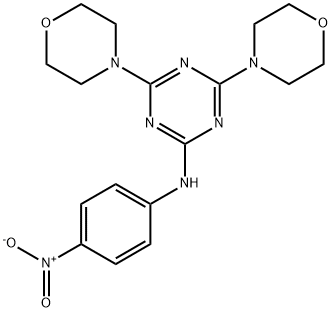
- Chemical Name:MHY1485
- CAS:326914-06-1
- MF:C17H21N7O4
- Structure:
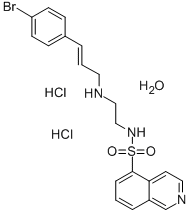
- Chemical Name:H-89 DIHYDROCHLORIDE HYDRATE
- CAS:130964-39-5
- MF:C20H20BrN3O2S.2ClH.H2O