Calicheamicin: Bioactivity, Mechanism of action, Application
General description
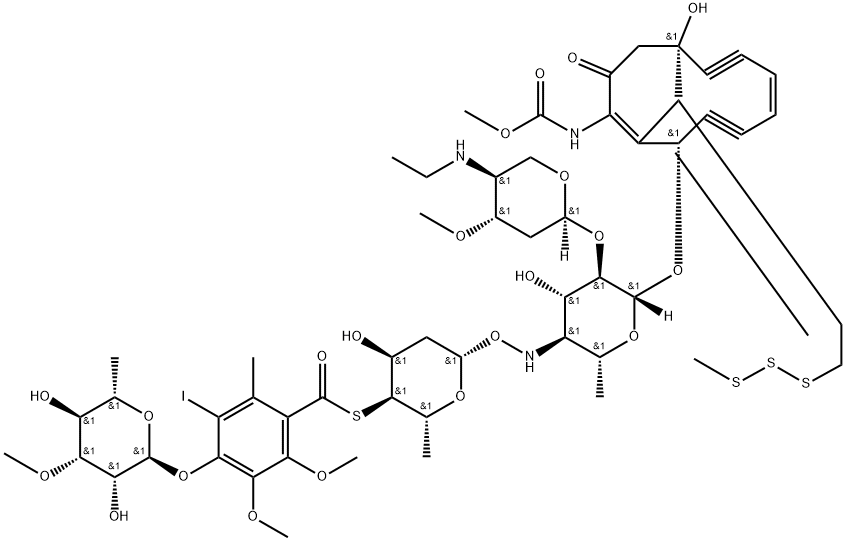
Calicheamicins (CLM) is an endiyne antitumor antibiotic isolated from the fermentation broth of the rare Micromonospora actinomycetes.
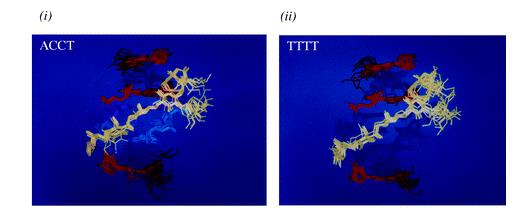
Figure 1 Overlays of 10 structures generated by simulated annealing for the (i) calicheamicin?ACCT and (ii) calicheamicin?TTTT complexes. Calculated RMSDs for the complexes, excluding the terminal base-pairs and sodium atoms, were 1.106 ? for ACCT and 1.135 ? for TTTT, respectively.
Bioactivity
Antitumor
It is assumed that the efficient antitumor activity of calicheamicin gamma1 is mediated by its ability to introduce DNA double-strand breaks in cellular DNA. To test this assumption we have compared calichearnicin gamma1-mediated cleavage of cellular DNA and purified plasmid DNA. Cleavage of purified plasmid DNA was not inhibited by excess tRNA or protein indicating that calicheamicin gamma1 specifically targets DNA. Cleavage of plasmid DNA was not affected by incubation temperature. In contrast, cleavage of cellular DNA was 45-fold less efficient at 0degreesC as compared to 37degrees due to poor cell permeability at low temperatures. The ratio of DNA double-strand breaks (DSB) to single-stranded breaks (SSB) in cellular DNA was 1:3, close to the 1:2 ratio observed when calichearnicin gamma1 cleaved purified plasmid DNA. DNA strand breaks introduced by calichearnicin gamma1 were evenly distributed in the cell population as measured by the comet assay. Calicheamicin gamma1-induced DSBs, were repaired slowly but completely and resulted in high levels of H2AX phosphorylation and efficient cell cycle arrest. In addition, the DSB-repair deficient cell line Mo59J was hyper sensitive to calicheamicin gamma. The data indicate that DSBs is the crucial damage after calichearnicin gamma1 and that calicheamicin gamma1-induced DSBs are recognized normally. The high DSB:SSB ratio, specificity for DNA and the even damage distribution makes calicheamicin gamma1 a superior drug for studies of the DSB-response and emphasizes its usefulness in treatment of malignant disease [1].
Calicheamicin gamma1 (Cal gamma1) is a hydrophobic enediyne antibiotic known to cleave the DNA and lead to apoptosis in a variety of cells. Herein, we show that Cal gammal exhibits a 1000-times stronger suppressogenic effect on antigen-specific (diabetogenic), and naive CD4 T cells than Doxorubicin (Dox), another strong apoptotic drug. The thymic precursors and mature T cells incubated with Cal gamma1 for only 30 min showed a drastic decrease or loss of cytokine production and proliferation following stimulation with the immunogenic peptide, or with CD3 and CD28 antibodies. The suppressogenicity of Cal gamma1 correlated with a rapid and non-selective degradation of RNA, whereas the DNA cleavage occurred at a later time point and at higher doses. Cal gamma1 may represent a potential therapeutic agent to eliminate self-reactive T cells in autoimrnune diseases, providing that is delivered by antigen-specific T-cell ligands. Targeting of highly suppressogenic drugs such as Cal gamma1 to autoreactive T cells may reduce considerable the therapeutic dose and the drug-related side effects [2].
Calicheamicin gamma1 (Cal gamma1) is a hydrophobic enediyne leading to cleavage of the DNA double stranded and apoptosis in a variety of cells. Herein, we provide evidence that Cal gamma1 induces cell growth arrest and suppresses the cytokine production in antigen-specific CD4 T-cells previously stimulated in vitro with the cognate immunogenic peptide. The cytotoxicity of Cal gamma1 relied more on the rate of cellular uptake than its chemical stability. Thus, the minimal time required for T-cells to uptake a lethal dose of Cal gamma1 in. culture was less than 30 min, whereas the drug toxicity was reduced by 80% within 2 h of incubation with culture medium alone. Our present and previous work indicate that Cal gamma1 is a powerful suppressogenic drug on antigen-specific T-cells. It also suggests that Cal gamma1 could be used as immunospecific therapeutic agent in autoimmune diseases to eliminate self reactive T-cells, providing that is delivered specifically throughout a T-cell ligand such as soluble peptide-MHC chimeras [3].
In this study we elucidated the role of ATP-binding cassette (ABC) multi-drug transporter proteins and cellular factors such as Bcl-2 expression and CD33 down-modulation contributing to free and hP67.6 mAb linked calicheamicin-gamma 1 (CalC-gamma 1) resistance. We analyzed in a well designed HL60 cell system the relationship between the expression of ABC proteins, Bcl-2 and CD33 modulation with the activity of free and mAb-linked CalC-gamma 1. The results herein reported and discussed, strongly suggest that both MDR1-Pgp and MRP1 efflux systems are engaged by CalC-gamma 1, but only MDR1-Pgp over-expression efficiently abrogates drug cytotoxicity in MDR cells. Paradoxically, Bcl-2 expression, as observed for other anticancer compounds belonging to the enediyne family of drugs, confers CalC-gamma 1 susceptibility rather than resistance in HL60 cells. Further, the isolation of a resistant HL60 subline (HL60AL) that was developed by exposing the parental sensitive cells to sub-effective doses of gemtuzumab ozogamicin (GO) over an extended period of time shows a reduced level of CD33 expression that represents an important escape mechanism of HL60 MDR cells to the cytotoxic effect of GO [4].
Application
Structural characterization of a calicheamicin DNA complex by NMR
The enediynes, including neocarzinostatin, calicheamicin, esperamicin, and dyneamicin, are an important class of antitumor antibiotics that cleave DNA. In spite of intense interest in the enediynes as potential drugs, there is no detailed structural information about how any of these compounds interacts with DNA. We report the first NMR studies of a complex between an enediyne, calicheamicin gamma1, and DNA. Calicheamicin gamma1 cleaves DNA in a double-stranded fashion at oligopyrimidine/oligopurine sequences. The molecular basis for the selective recognition of pyrimidine/purine runs is not well understood. Using NMR we have shown that calicheamicin gamma1 binds to the non-self-complementary DNA duplex, d[GTGACCTG]-d[CAGGTCAC], where ACCT is the recognition sequence. The DNA distorts upon binding to accommodate the drug. The distortion is largest at the CpC step of the recognition sequence and appears to be associated with a widening of the minor groove. A preliminary analysis of the data indicates that the drug itself does not distort much upon binding. It is proposed that binding selectivity reflects the ability of oligopyrimidine sequences to distort to accommodate the more rigid drug [5].
Interaction of calicheamicin gamma(I)(1) and its related carbohydrates with DNA-protein complexes
We report studies of the contribution of DNA structure, holding the sequence constant, to the affinity of calicheamicin gamma(1)(I) and its aryltetrasaccharide moiety for DNA. We used polynucleotide chains as models of known protein-binding sequences [the catabolite activator protein (CAP) consensus sequence, AP-1 and cAMP response element (CRE) sites] in their free and protein-bound forms. The proteins
You may like
Related articles And Qustion
See also
Lastest Price from Calicheamicin manufacturers
US $8.00-1.00/kg2024-04-05
- CAS:
- 108212-75-5
- Min. Order:
- 1kg
- Purity:
- 99%
- Supply Ability:
- g-kg-tons, free sample is available