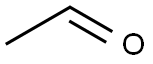
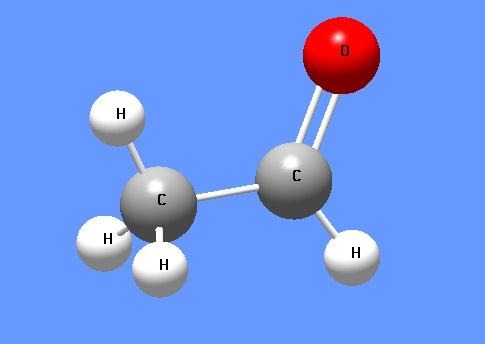
Acetaldehyde: Sources and Multiple DNA repair pathways
Sources of acetaldehyde
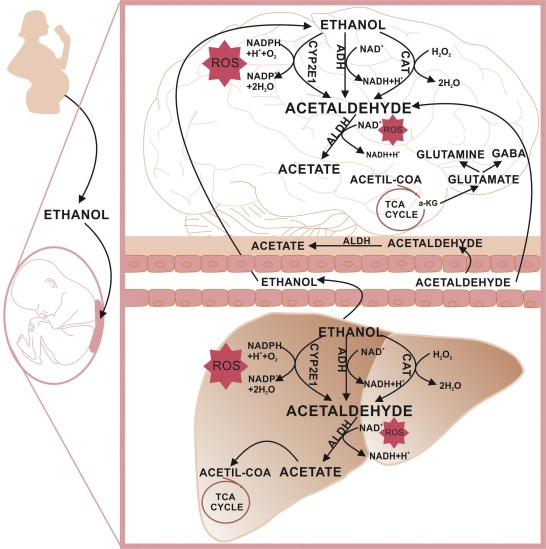
Acetaldehyde is the primary metabolite of alcohol and is present in many environmental sources including tobacco smoke. Ethanol is metabolized mainly by alcohol dehydrogenase (ADH) to produce acetaldehyde. At high levels of ethanol consumption, cytochrome P450 2E1 (CYP2E1) becomes involved in metabolizing ethanol to acetaldehyde. Catalase (CAT) metabolizes ~60% of ethanol within the brain where physiologically active ADH is lacking. Acetaldehyde is oxidized into acetate mainly by aldehyde dehydrogenase (ALDH). Acetate may then be converted into acetyl- Coenzyme A, which can be oxidized in the tricarboxylic acid cycle (TCA). The α- ketoglutarate from TCA may be used as a source of glutamate, glutamine, or GABA. Ethanol metabolism results in the formation of NADH and, thus, changes the cellular redox state. Re-oxidation of NADH via the mitochondrial electron transport chain results in the formation of reactive oxygen species (ROS).
Multiple DNA repair pathways prevent acetaldehyde-induced mutagenesis in yeast
Acetaldehyde is genotoxic, whereby it can form DNA adducts and lead to mutagenesis. Individuals with defects in acetaldehyde clearance pathways have increased susceptibility to alcohol-associated cancers. Moreover, a mutation signature specific to acetaldehyde exposure is widespread in alcohol and smoking-associated cancers. However, the pathways that repair acetaldehyde-induced DNA damage and thus prevent mutagenesis are vaguely understood. Here, we used Saccharomycescerevisiae to systematically delete genes in each of the major DNA repair pathways to identify those that alter acetaldehyde-induced mutagenesis. We found that deletion of the nucleotide excision repair (NER) genes, RAD1 or RAD14, led to an increase in mutagenesis upon acetaldehyde exposure. Acetaldehyde-induced mutations were dependent on translesion synthesis as well as DNA inter-strand crosslink (ICL) repair in Drad1 strains. Moreover, whole genome sequencing of the mutated isolates demonstrated an increase in C?A changes coupled with an enrichment of gCn?A changes in the acetaldehyde-treated Drad1 isolates. The gCn?A mutation signature has been shown to be diagnostic of acetaldehyde exposure in yeast and in human cancers. We also demonstrated that the deletion of the two DNA-protein crosslink (DPC) repair proteases, WSS1 and DDI1, also led to increased acetaldehyde-induced mutagenesis. Defects in base excision repair (BER) led to a mild increase in mutagenesis, while defects in mismatch repair (MMR), homologous recombination repair (HR) and post replicative repair pathways did not impact mutagenesis upon acetaldehyde exposure. Our results in yeast were further corroborated upon analysis of whole exome sequenced liver cancers, wherein, tumors with defects in ERCC1 and ERCC4 (NER), FANCD2 (ICL repair) or SPRTN (DPC repair) carried a higher gCn?A mutation load than tumors with no deleterious mutations in these genes. Our findings demonstrate that multiple DNA repair pathways protect against acetaldehyde-induced mutagenesis.
References:
[1] PORCHER L, VIJAYRAGHAVAN S, MCCOLLUM J, et al. Multiple DNA repair pathways prevent acetaldehyde-induced mutagenesis in yeast[J]. bioRxiv - Genetics, 2024. DOI:10.1101/2024.01.07.574575.
Related articles And Qustion
Lastest Price from Acetaldehyde manufacturers

US $1.00/KG2025-04-21
- CAS:
- 75-07-0
- Min. Order:
- 1KG
- Purity:
- 99%
- Supply Ability:
- 10 mt

US $9.00/KG2024-10-11
- CAS:
- 75-07-0
- Min. Order:
- 1KG
- Purity:
- 99.8%
- Supply Ability:
- 100tons