DNA damage
DNA damage refers to the changes of structure of the cell DNA in the body caused by the physical or chemical contamination factors around the environment. There are many types of chemicals can cause DNA damage , for example, nitrosamines firstly metabolize and break down in the body , resulting in an alkylating agent - diazoalkane,which leads to the DNA alkylation , causing the carrier of genetic information to produce irreparable changes and resulting in carcinogenic effects . Another example is Benzo (a) pyrene of polycyclic aromatic hydrocarbons (PAH) , which in mixed function oxidase activation,can form the compound having structure that may have an affinity for the electron - epoxide. Such epoxides can combine with DNA and other intracellular macromolecules nucleophilic groups ,which will cause DNA damage. When such damage can not be repaired or there are repair errors, it is possible to make cells cancerous. Some metals such as nickel, beryllium, chromium and other carcinogens, it is possible to form a stable complex with RNA complex residues, resulting in carcinogenesis.
DNA repair
DNA repair refers to the recovery phenomenon of structure after the DNA molecule inside living cells is damaged. It is the result of action of many enzymes within the cells of the body. In 1968, American scholar J • E • Cleaver first discovered that the autosomal recessive cancerous actinic disease in human ,which is xeroderma pigmentosum disease , is caused by excision and repair kinetic energy defect which results from a gene mutation D NA . The discovery not only provides an important molecular evidence for the mechanism of cancer , but also makes study about DNA damage and repair into the medical field. On the14th International Genetics Conference in 1979 held in Moscow DNA damage and repair is listed as one of the important issues, the DNA repair research has become an important research topic of molecule genetics and medical genetics and oncology.
There are several types of DNA molecular damage , such as the formation of thymine dimers which is the major manner that UV damages DNA molecules ; x-ray, the r-ray irradiating cell, can make hydrogen bonds between double-stranded DNA molecule break , it can also cause single-stranded or double-strand breaks; mitomycin C can cause cross-linking between single-stranded DNA molecule ,cross-linked chains of DNA molecules can cause breakage; DNA molecules can also occur individual base or nucleotide or the like change. DNA damage repair methods include optical resurrection, excision repair, repair after replication , adaptive restoration, repair of strand breaks, cross links repair. Studys on DNA damage repair mechanisms can help to understand gene mutations, cancer and aging reasons, and can also be applied to detect cancer-causing environmental factors.
- Structure:
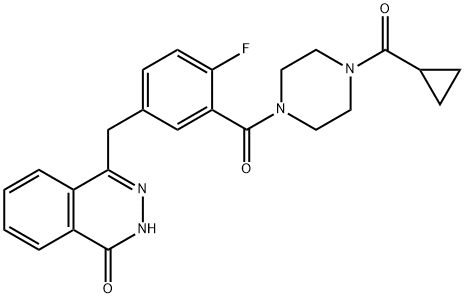
- Chemical Name:Olaparib
- CAS:763113-22-0
- MF:C24H23FN4O3
- Structure:
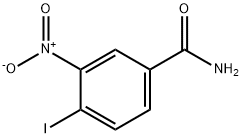
- Chemical Name:4-iodo-3-nitrobenzamide
- CAS:160003-66-7
- MF:C7H5IN2O3
- Structure:
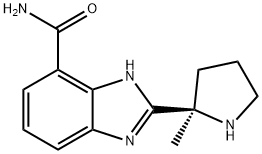
- Chemical Name:Veliparib
- CAS:912444-00-9
- MF:C13H16N4O
- Structure:
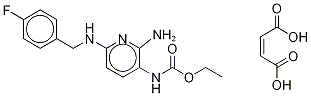
- Chemical Name:Flupirtine maleate
- CAS:75507-68-5
- MF:C15H17FN4O2.C4H4O4
- Structure:
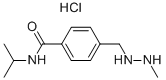
- Chemical Name:Procarbazine hydrochloride
- CAS:366-70-1
- MF:C12H20ClN3O
- Structure:
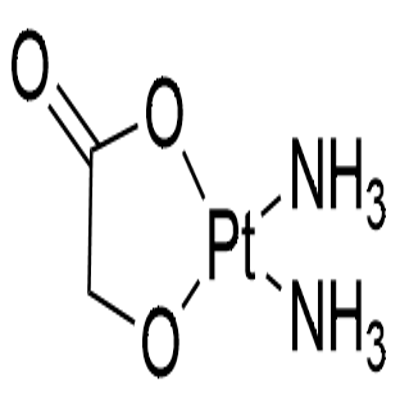
- Chemical Name:Nedaplatin
- CAS:95734-82-0
- MF:C2H6N2O3Pt
- Structure:
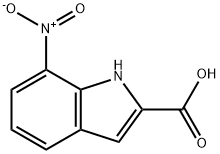
- Chemical Name:7-Nitroindole-2-carboxylic acid
- CAS:6960-45-8
- MF:C9H6N2O4
- Structure:
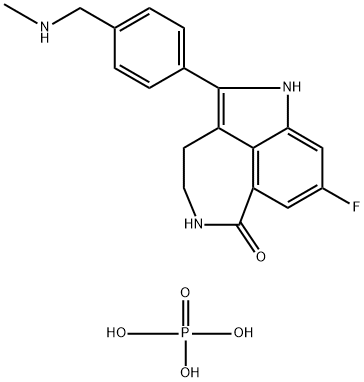
- Chemical Name:PF-01367338
- CAS:459868-92-9
- MF:C19H21FN3O5P
- Structure:
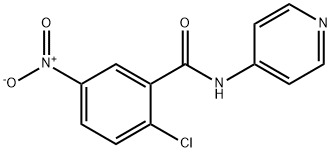
- Chemical Name:T0070907
- CAS:313516-66-4
- MF:C12H8ClN3O3
- Structure:
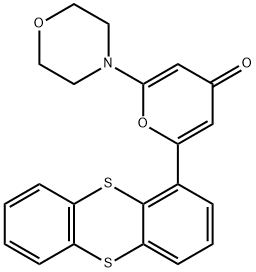
- Chemical Name:KU-55933
- CAS:587871-26-9
- MF:C21H17NO3S2
- Structure:
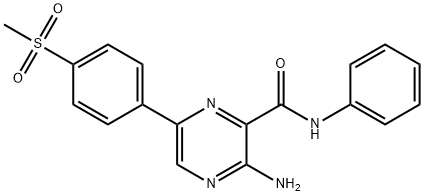
- Chemical Name:3-Amino-6-[4-(methylsulfonyl)phenyl]-N-phenyl-2-pyrazinecarboxamide
- CAS:1232410-49-9
- MF:C18H16N4O3S
- Structure:
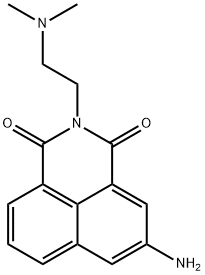
- Chemical Name:Amonafide
- CAS:69408-81-7
- MF:C16H17N3O2
- Structure:
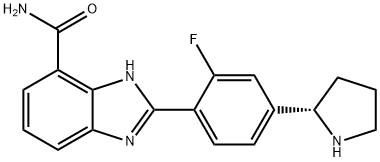
- Chemical Name:A-966492
- CAS:934162-61-5
- MF:C18H17FN4O
- Structure:
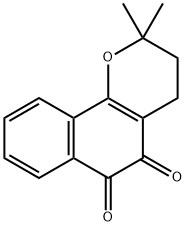
- Chemical Name:BETA-LAPACHONE
- CAS:4707-32-8
- MF:C15H14O3
- Structure:
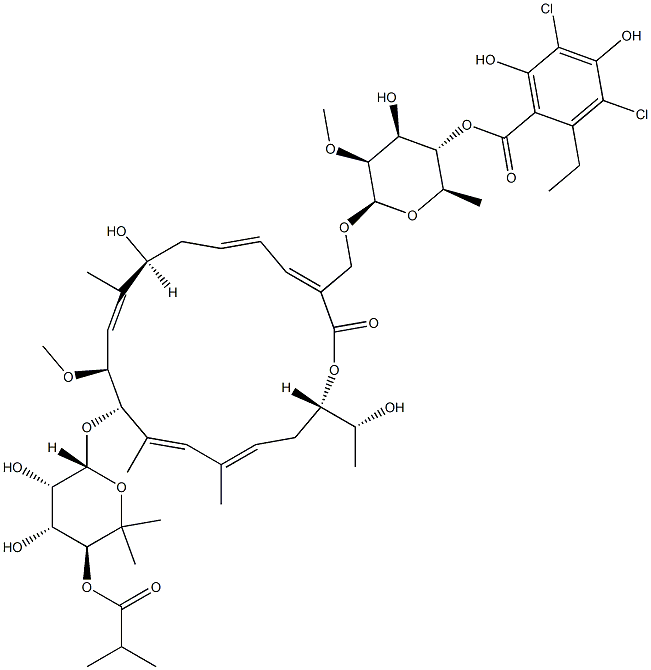
- Chemical Name:FidaxoMicin
- CAS:873857-62-6
- MF:C52H74Cl2O18
- Structure:
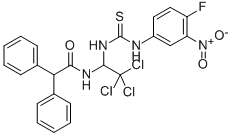
- Chemical Name:2,2-DIPHENYL-N-(2,2,2-TRICHLORO-1-[3-(4-FLUORO-3-NITROPHENYL)THIOUREIDO]ETHYL)ACETAMIDE
- CAS:905973-89-9
- MF:C23H18Cl3FN4O3S
- Structure:
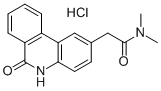
- Chemical Name:PJ-34
- CAS:344458-15-7
- MF:C17H17N3O2HCl
- Structure:
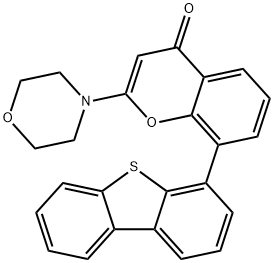
- Chemical Name:KU57788(NU7441)
- CAS:503468-95-9
- MF:C25H19NO3S
- Structure:
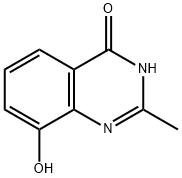
- Chemical Name:NU1025
- CAS:90417-38-2
- MF:C9H8N2O2
- Structure:
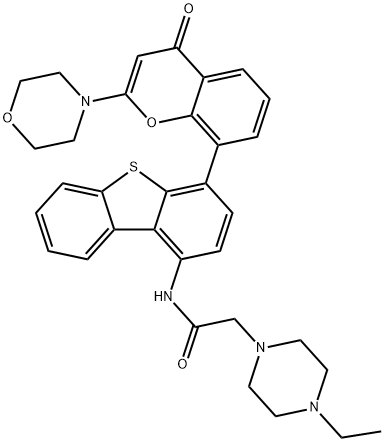
- Chemical Name:KU 0060648
- CAS:881375-00-4
- MF:C33H34N4O4S
- Structure:
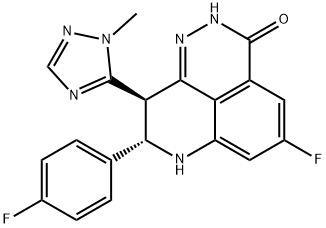
- Chemical Name:BMN 673
- CAS:1207456-01-6
- MF:C19H14F2N6O
- Structure:
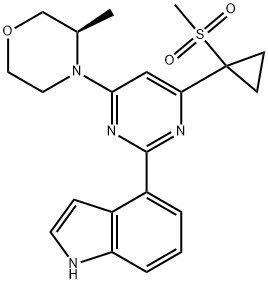
- Chemical Name:AZ20
- CAS:1233339-22-4
- MF:C21H24N4O3S
- Structure:
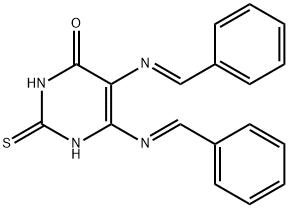
- Chemical Name:SCR7
- CAS:1533426-72-0
- MF:C18H14N4OS
- Structure:

- Chemical Name:PJ-34
- CAS:344458-19-1
- MF:C17H17N3O2
- Structure:
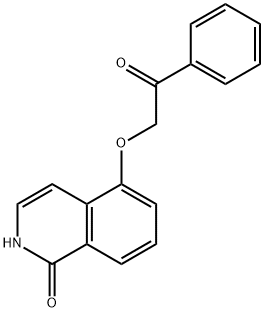
- Chemical Name:5-(2-Oxo-2-phenylethoxy)-3,4-dihydroisoquinolin-1(2H)-one
- CAS:1048371-03-4
- MF:C17H13NO3
- Structure:
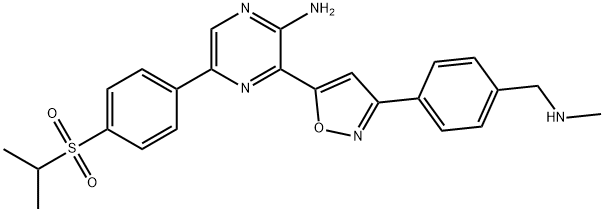
- Chemical Name:VE 822
- CAS:1232416-25-9
- MF:C24H25N5O3S
- Structure:
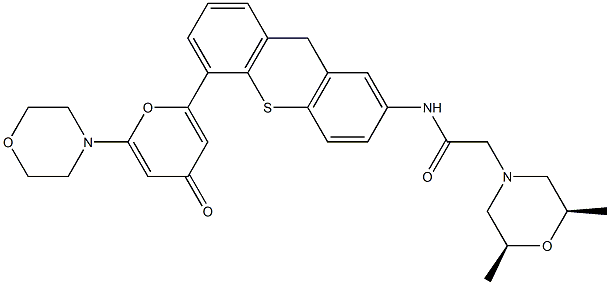
- Chemical Name:KU-60019
- CAS:925701-49-1
- MF:C30H33N3O5S
- Structure:
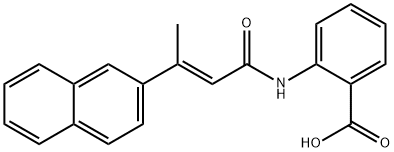
- Chemical Name:2-[[(2E)-3-(2-NAPHTHALENYL)-1-OXO-2-BUTEN-1-YL]AMINO]BENZOIC ACID
- CAS:321674-73-1
- MF:C21H17NO3
- Structure:

- Chemical Name:CX-5461
- CAS:1138549-36-6
- MF:C27H27N7O2S
- Structure:
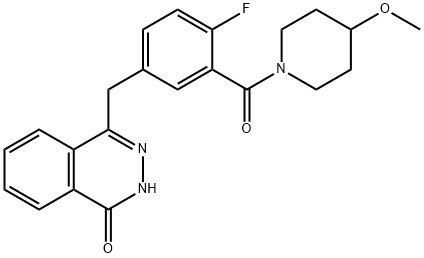
- Chemical Name:AZD2461
- CAS:1174043-16-3
- MF:C22H22FN3O3
- Structure:
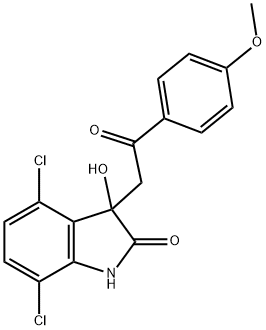
- Chemical Name:YK-4-279
- CAS:1037184-44-3
- MF:C17H13Cl2NO4
- Structure:
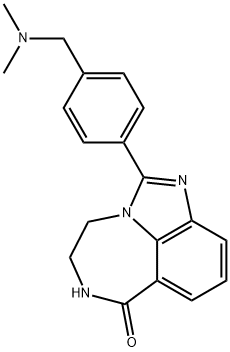
- Chemical Name:Imidazo[4,5,1-jk][1,4]benzodiazepin-7(4H)-one, 2-[4-[(dimethylamino)methyl]phenyl]-5,6-dihydro-
- CAS:328543-09-5
- MF:C19H20N4O
- Structure:
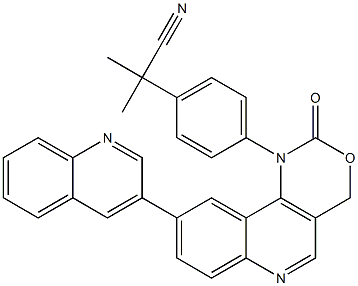
- Chemical Name:ETP46464
- CAS:1345675-02-6
- MF:C30H22N4O2
- Structure:
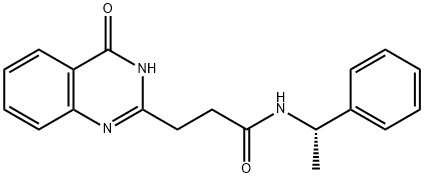
- Chemical Name:ME0328
- CAS:1445251-22-8
- MF:C19H19N3O2
- Structure:

- Chemical Name:BMH21
- CAS:896705-16-1
- MF:C21H20N4O2
- Structure:
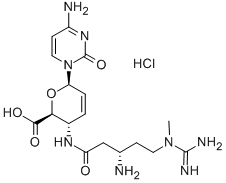
- Chemical Name:BLASTICIDIN S HYDROCHLORIDE
- CAS:3513-03-9
- MF:C17H27ClN8O5
- Structure:
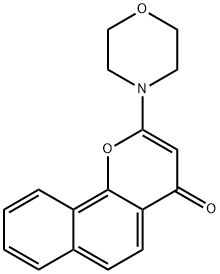
- Chemical Name:NU7026
- CAS:154447-35-5
- MF:C17H15NO3
- Structure:
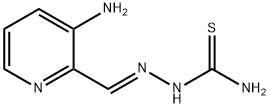
- Chemical Name:Triapine
- CAS:200933-27-3
- MF:C7H9N5S
- Structure:
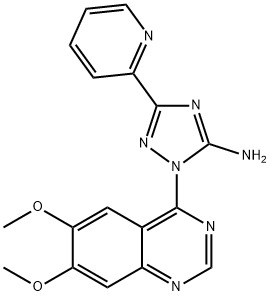
- Chemical Name:CP466722
- CAS:1080622-86-1
- MF:C17H15N7O2
- Structure:
- Chemical Name:Voreloxin
- CAS:175414-77-4
- MF:C18H19N5O4S