New Study Reveals Genetic Polymorphisms Affecting Metformin Efficacy
General Description
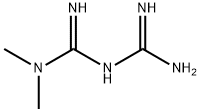
Metformin is a hydrophilic drug that enters cells through active transporters, including PMAT, OCT1, OCT3, and OCT2. It is primarily absorbed by the small intestine and distributed to various tissues, with higher concentrations in the liver due to direct blood supply from the portal vein. Metformin's pharmacokinetics inform its dosing regimen for managing diabetes. Pharmacogenomics research has identified genetic polymorphisms in cation transporters that affect metformin's clinical effectiveness and pharmacokinetics. Variants in SLC22A1 are associated with reduced hepatic uptake and impaired glucose-lowering effects, while variants in SLC47A1 are linked to enhanced renal clearance and improved glycemic control. Further studies are needed to fully elucidate genetic influences on metformin's clinical response.

Figure 1. Tablets of metformin
Metformin Administration and Plasma Concentrations
Metformin is a commonly used medication for diabetic patients. After oral administration, metformin is partially absorbed by the small intestine and quickly distributed to various tissues, while the concentration in the gastrointestinal tract remains high. Peak plasma concentration is reached within 3 hours, with levels increasing from 1.0 to 1.6 mg/ml (about 6 to 10 mM) for a 0.5 g dose, and up to 3 mg/ml (about 18 mM) for a 1.5 g dose. The average half-life of metformin in plasma is approximately 20 hours. In mice, when the human equivalent dose of 20 mg/kg/day is administered orally, plasma concentrations of up to 1.7 mg/ml (about 10 mM) are achieved. Biodistribution studies using labeled metformin in mice have shown accumulation in the gastrointestinal tract, kidney, and liver. It's worth noting that the liver receives a higher concentration of metformin compared to other organs due to direct blood supply from the portal vein. Liver concentrations of metformin can exceed 180 mmol/kg wet weight in normal rodents and 250 mmol/kg wet weight in diabetic rodents after a single dose of 50 mg/kg. Overall, understanding the pharmacokinetics of metformin is important in optimizing its dosing regimen and ensuring its efficacy in managing diabetes. 1
Cellular uptake
Metformin is a hydrophilic drug that exists in a positively charged form in the body. Due to its physicochemical properties, it cannot easily diffuse through cell membranes via passive diffusion. Instead, metformin enters cells through an active uptake process involving specific transporters. In the intestines, metformin is primarily absorbed by the plasma membrane monoamine transporter (PMAT) located on the luminal side of enterocytes. Organic cation transporter 1 (Oct1) on the basolateral membrane of enterocytes may facilitate metformin transport into the interstitial fluid. In the liver, metformin uptake is mainly mediated by OCT1 and possibly OCT3, both expressed on the basolateral membrane of hepatocytes. Renal elimination plays a crucial role in metformin clearance. In the kidneys, metformin is taken up into renal epithelial cells by OCT2 on the basolateral membrane. It is then excreted into urine through multidrug and toxin extrusion transporters MATE1 and MATE2. Metformin's cellular uptake involves these specific transporters, ensuring its distribution and elimination in the body. 2
Genetic Variability and Metformin Response
Pharmacogenomics research has revealed significant variability in both the clinical effectiveness and pharmacokinetics of metformin among diabetic patients, which can be attributed to genetic polymorphisms in cation transporters. Variants in the OCT1 gene SLC22A1 have been associated with reduced hepatic metformin uptake, resulting in impaired glucose-lowering effects. However, long-term observational studies have not consistently confirmed these findings. Conversely, variants in the MATE1 gene SLC47A1 have been linked to enhanced metformin efficacy in improving glycated hemoglobin (HbA1c) and glucose tolerance, possibly through increased renal clearance of the drug. Furthermore, new candidate genetic determinants, including single nucleotide polymorphisms in AMPK subunit genes and the LKB1 gene STK11, have been identified, along with a chromosome 11 locus encompassing several genes associated with glycemic variability in response to metformin therapy. The ataxia telangiectasia mutated (ATM) gene within this locus has been suggested as a potential candidate due to its association with insulin resistance and type 2 diabetes (T2D). Nevertheless, further research is necessary to fully elucidate genetic influences on the clinical response to metformin. 3
Reference
1. Tucker, G.T., Casey, C., Phillips, P.J., Connor, H., Ward, J.D., and Woods, H.F. (1981). Metformin kinetics in healthy subjects and in patients with diabetes mellitus. Br. J. Clin. Pharmacol. 12, 235–246.
2. Gong, L., Goswami, S., Giacomini, K.M., Altman, R.B., and Klein, T.E. (2012). Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet. Genomics 22, 820–827.
3. Shu, Y., Sheardown, S.A., Brown, C., Owen, R.P., Zhang, S., Castro, R.A., Ianculescu, A.G., Yue, L., Lo, J.C., Burchard, E.G., et al (2007). Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 117, 1422–1431.
You may like
Related articles And Qustion
Lastest Price from Metformin manufacturers

US $160.00-45.00/kg2025-04-21
- CAS:
- 657-24-9
- Min. Order:
- 1kg
- Purity:
- 99% purity,WhatsApp+86 18102676775
- Supply Ability:
- 20 tons

US $150.00-45.00/kg2025-04-21
- CAS:
- 657-24-9
- Min. Order:
- 1kg
- Purity:
- 99% Purity (What/sapp: +86 18145728414)
- Supply Ability:
- 1000 Tons/Month