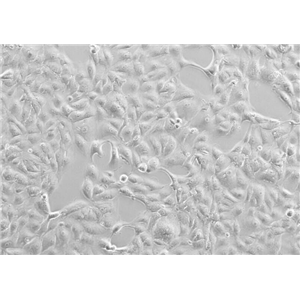
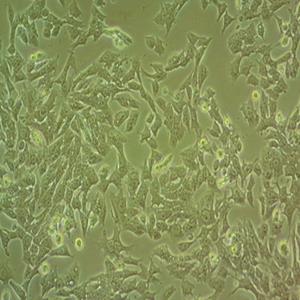
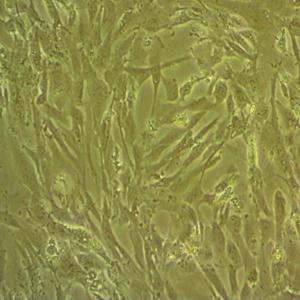
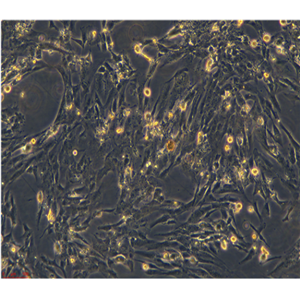
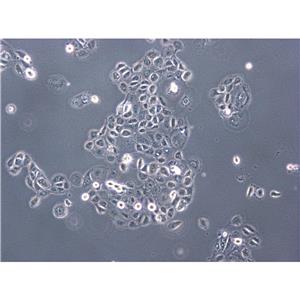
"NAMALWA人Burkitt's淋巴瘤细胞全年复苏|已有STR图谱
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:悬浮生长
细胞系的选择需要考虑到细胞系的功能特点、生长速率、铺板效率、生长条件和生长特征、克隆效率、培养方式等因素,如果您想高产量表达重组蛋白,您可以选择可以悬浮生长的快速生长细胞系。细胞培养的操作步骤主要包括传代、换液、冻存和复苏。这些步骤确保了细胞能够在实验室环境中长期存活并继续增殖。传代是将细胞从一个容器转移到另一个容器的过程,以扩大细胞数量;换液是为了清除代谢废物并补充新鲜培养基;冻存则是为了长期保存细胞,而复苏则是重新激活冷冻保存的细胞使其恢复正常生长。
换液周期:每周2-3次
HFL 1 Cells;背景说明:详见相关文献介绍;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:MIA Paca2细胞、Panc 04.03细胞、HCC-1937细胞
L929(NCTC) Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:C-26细胞、GP-293细胞、AHH1细胞
D10G41 Cells;背景说明:T淋巴细胞;AKR/J;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:SUM-102细胞、AR4IP细胞、bEnd3细胞
背景信息:最初是从一名患有Burkitt's淋巴瘤的患者的肿瘤组织中分离出来的。是一种高度恶性的B细胞淋巴瘤,常见于儿童和年轻人。有EB病毒基因组。
NAMALWA人Burkitt's淋巴瘤细胞全年复苏|已有STR图谱
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
贴壁细胞的传代培养,详细步骤如下:首先倒掉培养基,在这一步骤可以收集一些细胞上清做支原体检测;加入胰蛋白酶,一般T25是加2mL,盖好瓶盖,摇晃T25培养瓶,使胰蛋白酶均匀覆盖在细胞表面,放入培养箱2-3min,期间可在显微镜下观察,看到大部分细胞变圆,即可放入超净台,加入2倍的完全培养基,这里就是加4mL培养基,终止消化;将含有胰蛋白酶,细胞和培养基一起转移到离心管中,1000rpm/3min离心,去掉上清;新鲜的完全培养基重悬,根据细胞的生长特性和后续的实验需求进行传代,比如我养的Hepa1-6就长的比较快,不是着急用的话,我就会按1E6个细胞/T75培养瓶进行传代;但如果后两天要用,就会适当多传一点;还可通过显微镜计数后,直接用于细胞铺板,继续后续的实验。
Tissue Culture-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:CA-46细胞、HCC1419细胞、WC00059细胞
KBM-5 Cells;背景说明:慢性髓白血病;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:BPH-1细胞、University of Michigan-Urothelial Carcinoma-1细胞、Oregon J-111细胞
SH-SY5Y Cells;背景说明:据报道,该细胞的密度可高达1×106cells/cm2,具有中等水平的多巴胺β羟化酶的活性。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HMEC1细胞、COR L279细胞、ES-2细胞
AJ60 Cells(提供STR鉴定图谱)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
物种来源:人源、鼠源等其它物种来源
NAMALWA人Burkitt's淋巴瘤细胞全年复苏|已有STR图谱
形态特性:淋巴母细胞样
贴壁细胞消化传代时通常采用两种方法:一、加入胰酶等细胞脱落后,再加培养基中止胰酶作用,离心传代;二、加入胰酶后,镜下观察待细胞始脱落时,弃胰酶,加培养分瓶。但前者太麻烦,而后者有可能对细胞施加胰酶选择,因为总是贴壁不牢的细胞先脱落,对肿瘤细胞来说,这部分细胞有可能是恶性程度较GAO的细胞亚群。一种简单的消化传代方法。加入PBS洗去血清或加入胰酶先中和血清的作用(30s),弃之,再加入适量胰酶作用10s-40s(根据细胞消化的难易程度),弃之,这样依赖残余的胰酶就可将细胞消化单细胞。对于较难消化的细胞,可以用2%利多卡因消化5-8分钟,然后再弃去,加培养基吹打也可以,对细胞的影响不大。不用PBS也不用Hanks洗,只要把旧培养吸的干净一点,直接加酶消化应该不会有什么问题。弃培养后,用0.04%的EDA冲洗一次,再用1/4v的0.04%的EDA室温孵育5min,弃取大部分EDA,加入与剩余EDA等量的胰酶(预热)总体积1/10v。消化到有细胞脱落。不过有人说EDA对细胞不HAO,有证据吗?培养的BASMC:倒掉旧培养加入少量胰酶冲一下,倒掉再加入0.125-0.25%胰酶约6-10滴或1ml(25ml bole)消化再加入适量新培养基中和,并分瓶这种方法简单、省事;效果很HAO并且不损失细胞!
INS-1 Cells;背景说明:该细胞源自X射线照射的移植胰岛瘤的大鼠,胰岛素阳性,可合成胰岛素原I和II,可用于beta细胞功能研究。;传代方法:1:3—1:6传代,每周换液2—3次;生长特性:贴壁生长;形态特性:不规则,多角;相关产品有:BALL1细胞、22Rv1细胞、COLO 206细胞
RPMI8226/S Cells;背景说明:来源于一位61岁的男性浆细胞瘤患者;可产生免疫球蛋白轻链,未检测到重链。;传代方法:按1:2传代,5-6小时可以看到细胞分裂;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:HFL-1细胞、SMA-560细胞、BEL7404细胞
Hepa-RG Cells;背景说明:肝癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SF 763细胞、DHL4细胞、NCIH1373细胞
CCD 966 SK Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:A2780 CP70细胞、149PT细胞、BE2-C细胞
H522 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HUASMC细胞、JHH-2细胞、Daudi细胞
PLMVEC Cells;背景说明:肺微血管;内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:JB6 clone 30, subclone 7b细胞、HTh-74细胞、L(TK-)细胞
Tadarida brasiliensis 1 lung Cells;背景说明:肺;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:JCA-1细胞、SCC15细胞、P30/OHK细胞
P30/OHK Cells;背景说明:详见相关文献介绍;传代方法:10^5 cells/60mm dish;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:WSU-HN13细胞、LCD细胞、PAEC细胞
M619 Cells;背景说明:脉络膜黑色素瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H498细胞、PC-10细胞、UM-RC-2细胞
C17.2 Cells;背景说明:神经干细胞;C57BL/6 x CD-1;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:U 138 MG细胞、Eca109细胞、U266S细胞
H-2228 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,每周2-3次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:VB细胞、KE-39细胞、KP-N-RT-BM细胞
SNU-638 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Hu-P-T4细胞、VeroC1008细胞、Anip[973]细胞
DSL6A/C1 Cells;背景说明:胰腺癌;雄性;Lewis;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HSC-4细胞、SW 900细胞、LK-2细胞
hESC Cells;背景说明:子宫内膜;间质细胞;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:BCAP37细胞、TE12细胞、174xCEM.T2细胞
CNE2Z Cells;背景说明:鼻咽癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:LOU-NH-91细胞、COS-1细胞、CNE细胞
138MG Cells;背景说明:星形细胞瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:TYKnu细胞、HEK293-EBNA1细胞、DrG细胞
HEK293-EBNA Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:10传代;每周2次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:Panc 2.03细胞、Hs746T细胞、U-937细胞
Abcam A-549 TMEM64 KO 2 Cells(提供STR鉴定图谱)
AG02655 Cells(提供STR鉴定图谱)
BayGenomics ES cell line CSH727 Cells(提供STR鉴定图谱)
BayGenomics ES cell line RST011 Cells(提供STR鉴定图谱)
BEAS-2B/EJ-ras Cells(提供STR鉴定图谱)
CHO-MCHR2 Cells(提供STR鉴定图谱)
DA02876 Cells(提供STR鉴定图谱)
ES[MC1R(20):tetJarid2(37)] Cells(提供STR鉴定图谱)
GM06548 Cells(提供STR鉴定图谱)
SK HEP 01 Cells;背景说明:SK-HEP-1细胞系已被鉴定为内皮来源。该细胞系为异倍体女性人(XX),染色体在亚三倍体范围内。在裸鼠中,它能形成与肝癌相一致的大细胞癌;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HcaF细胞、Wien-133细胞、CAMA-1细胞
T-47-D Cells;背景说明:浸润性导管癌;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:BTI-TN5B1-4细胞、Panc_08_13细胞、KMST6细胞
Hs 746.T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SK_N_BE2C细胞、U87 MG细胞、Pt K2 (NBL-5)细胞
NMCG-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:GM00215细胞、TE-32细胞、CAL 62细胞
CV 1 Cells;背景说明:CV-1细胞株是1964年由JensenFC等建系的,源自成年雄性非洲绿猴肾,被用于Rous肉瘤病毒的转染研究。可作为SV40载体的转染宿主。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:hADSCs细胞、BALL-1细胞、Z310细胞
Hs821T Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;每周换液2-3次;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:ATC241细胞、HCC2279细胞、MZ-CRC-1细胞
OCILY3 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:Madison细胞、mouse Inner Medullary Collecting Duct-3细胞、HEK AD293细胞
Mouse Bladder Tumor line-2 Cells;背景说明:膀胱移行细胞癌;C3H/He;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NPA87-1细胞、Porcine Kidney-15细胞、IPLB-SF-21细胞
NAMALWA人Burkitt's淋巴瘤细胞全年复苏|已有STR图谱
E304 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SR-786细胞、3LL细胞、U138细胞
WM115 Cells;背景说明:黑色素瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NE1细胞、SN12PM6细胞、Jurkat FHCRC细胞
Mv1Lu Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:P3X63-Ag8.653细胞、KU-812细胞、BIU-87细胞
MDA-MB-435-S Cells;背景说明:MDA-MB-435S是一种纺锤形的细胞,1976年由其亲本(435)中筛选得到。435是从31岁的转移性乳腺导管腺癌女性患者胸水中分离得到。当用荧光染料对微管蛋白进行染色时亲本细胞显现散布特征(II型)。最近通过cDNA阵列研究表明,亲本(MDA-MB-435)可归入黑素瘤起源。;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:纺锤形;相关产品有:Kit225-K6细胞、NTERA2D1细胞、Hs 737.T细胞
PA I Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:10传代;每周2-3次。 ;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:GM00637H细胞、P3J.HR1K细胞、EU-2细胞
MDA-MB-157 Cells;背景说明:该细胞源自一位患有乳腺髓样癌的44岁黑人女性,表达WNT7B癌基因,细胞与细胞边界处有细胞桥粒、微绒毛、张力细丝。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:A 172细胞、KGN细胞、LOUNH91细胞
LS 513 Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:4传代,每周换液2次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:H292细胞、Molm14细胞、XuanWei Lung Cancer-05细胞
GM11085 Cells(提供STR鉴定图谱)
HAP1 CMTR1 (-) 2 Cells(提供STR鉴定图谱)
Hs 888Lu Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:Ramos 2G6 4C10细胞、KP2细胞、KYSE70细胞
RCC4 Cells;背景说明:肾透明细胞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:KM933细胞、NCI-SNU-423细胞、HEK 293-F细胞
RPMI No. 1846 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H2081细胞、MDA-MB-231 #4175细胞、SK-OV-3细胞
HOP 92 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:WiDr/S细胞、624mel细胞、DI-TNC1细胞
ID8 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:IPI-2I细胞、RA 1细胞、FHs 74 Int细胞
B16-F0 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Hs737T细胞、AK细胞、HOEC细胞
H.Ep.-2 Cells;背景说明:最初认为这个细胞源自喉上皮癌,但随后通过同功酶分析、HeLa标记染色体和DNA指纹分析发现,起源细胞已被HeLa污染。 角蛋白免疫过氧化物酶染色阳性。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:SNU-886细胞、PLA802细胞、BT-B细胞
H69/P Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:4传代,每周换液2次;生长特性:悬浮生长,聚团;形态特性:聚团悬浮;相关产品有:Mo 59J细胞、SNU-1040细胞、HFT-8810细胞
HG1943 Cells(提供STR鉴定图谱)
IMR-32rDOX20 Cells(提供STR鉴定图谱)
LXKC40 Cells(提供STR鉴定图谱)
ND04385 Cells(提供STR鉴定图谱)
PeCa-UkHb-01 Cells(提供STR鉴定图谱)
Ubigene HeLa LIFR KO Cells(提供STR鉴定图谱)
XLC050 Cells(提供STR鉴定图谱)
HF2303 Cells(提供STR鉴定图谱)
SU86-86 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:H-4细胞、S3 HeLa细胞、UPCI:SCC090细胞
NUGC-4 Cells;背景说明:胃癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:半贴壁;形态特性:详见产品说明书;相关产品有:WSU-DLCL-2细胞、CF-1 MEF细胞、Be-Wo细胞
CORL105 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HT-29细胞、Ontario Cancer Institute-Acute Myeloid Leukemia-3细胞、MDA-MB 361细胞
LK 2 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MZ-CRC-1细胞、LADMAC细胞、J774 A1细胞
CaSki Cells;背景说明:这株细胞是从小肠肠系膜转移灶的细胞中建立的。 据报道,它含有完整的HPV-16(每个细胞大约600个拷贝)和HPV-18相关序列。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:HSF2细胞、EL 4细胞、KMS26细胞
CaSki Cells;背景说明:这株细胞是从小肠肠系膜转移灶的细胞中建立的。 据报道,它含有完整的HPV-16(每个细胞大约600个拷贝)和HPV-18相关序列。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:HSF2细胞、EL 4细胞、KMS26细胞
HL 60 Cells;背景说明:该细胞由CollinsSJ从一位患有急性早幼粒细胞性白血病的36岁白人女性的外周血中分离建立;可自发分化,或在盐、次黄嘌呤、佛波醇肉豆蔻酸(PMA,TPA)、DMSO(1%to1.5%)、D和视黄酸的刺激下发生分化;PMA刺激后可分泌TNF-α。该细胞具有吞噬活性和趋化反应,癌基因myc阳性,表达补体受体和FcR。;传代方法:维持细胞浓度在1×105-1×106/ml,每2-3天换液1次。;生长特性:悬浮生长;形态特性:髓母细胞样;相关产品有:SW 948细胞、B-104细胞、RERFLCMS细胞
MAEC Cells;背景说明:主动脉;内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NCIH820细胞、TF1细胞、RBL细胞
IGROV1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI.H226细胞、GH3细胞、SW982细胞
NR 8383 Cells;背景说明:NR8383(正常大鼠,1983年8月3日)来源于肺灌洗时的正常大鼠肺泡巨噬细胞。细胞在gerbil肺细胞连续培养液存在下培养了大约8-9个月。随后,不再需要外源生长因子。通过有限稀释法从单个细胞克隆并亚克隆NR8383细胞,并三次用软琼脂亚克隆。细胞表现出巨噬细胞的特性,吞噬酵母多糖和铜绿,非特异性脂酶活性,Fc受体,氧化降解;分泌IL-1,TNFbeta和IL-6,可重复地响应外源生长因子。NR8383细胞响应博莱霉素,分泌TGFbeta前体。在博莱霉素刺激下,TGFbe;传代方法:1:2传代;生长特性:半贴壁生长;形态特性:巨噬细胞;相关产品有:RH-30细胞、SF 539细胞、PLC8024细胞
CCC-HHM-2 Cells;背景说明:心肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CL40细胞、NCIH2171细胞、CCD19-Lu细胞
CCC-HEL-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SF763细胞、MDA.MB.231细胞、WM-266-mel细胞
H-596 Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:8传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:H-661细胞、CAL62细胞、Tb1Lu细胞
COLO684 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:GP5d细胞、MDA134细胞、SNUC1细胞
Line 697 Cells;背景说明:B淋巴细胞白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:HUT 226细胞、L-cells细胞、HeLa-S3细胞
SCVIi105-A Cells(提供STR鉴定图谱)
H-1522 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KNS-42细胞、CAL-78细胞、Hs 766.T细胞
HcerEpic Cells;背景说明:子宫颈;上皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-596细胞、Hepa1-6细胞、hUC-MSC细胞
COLO 320 DM Cells;背景说明:该细胞可产生5-羟色胺、去甲、、ACTH和甲状旁腺素。角蛋白、波形蛋白弱阳性。培养条件: RPMI 1640 10%FBS;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮+贴壁;形态特性:淋巴细胞;相关产品有:HT 1197.T细胞、WPMY1细胞、SK-MEL-2-LUC细胞
RS4:11 Cells;背景说明:详见相关文献介绍;传代方法:每周2-3次;生长特性:悬浮生长;形态特性:成淋巴细胞;相关产品有:PC-3/M细胞、Ramos (RA 1)细胞、COLO-394细胞
RGE Cells;背景说明:肾小球;内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs 683.T细胞、L615细胞、NOZC-1细胞
HSAEC1-KT Cells;背景说明:肺;HGNC-TERT转化;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SW-403细胞、Duke University 4475细胞、H-1623细胞
LLCMK2 Cells;背景说明:胚胎;肾;自发永生;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:L615细胞、Hs888Lu细胞、Hs_578t细胞
NCI-H82 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:5传代,每周换液2-3次;生长特性:悬浮生长;形态特性:上皮细胞;相关产品有:VSC4.1细胞、PT67细胞、H2108细胞
NAMALWA人Burkitt's淋巴瘤细胞全年复苏|已有STR图谱
Hi-5 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HCC1438细胞、SNU-C2A细胞、As-PC1细胞
H-2195 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H-226细胞、TALL1细胞、MV 4;11细胞
EFM-192A Cells;背景说明:乳腺癌;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H524细胞、C3H10T1/2 clone8细胞、ZYM-SVEC01细胞
OSC-19 Cells;背景说明:舌鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:BE2_C细胞、HNE-3细胞、HCEpiC细胞
NT2D1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:EJ-1细胞、OCI-Ly10细胞、H1755细胞
RC-4B/C Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-HUT-520细胞、Monomac-1细胞、SHEP1细胞
BayGenomics ES cell line RRI002 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XH413 Cells(提供STR鉴定图谱)
ES[MC1R(20):tetSub1(12)] Cells(提供STR鉴定图谱)
NAL1A clone C4E10 Cells(提供STR鉴定图谱)
UC8-1B9 Cells(提供STR鉴定图谱)
MC-B11/14 Cells(提供STR鉴定图谱)
" "PubMed=170370; DOI=10.1099/0022-1317-28-2-207
Adams A., Strander H., Cantell K.
Sensitivity of the Epstein-Barr virus transformed human lymphoid cell lines to interferon.
J. Gen. Virol. 28:207-217(1975)
PubMed=216485
Higgins N.P., Strauss B.S.
Differences in the ability of human lymphoblastoid lines to exclude bromodeoxyuridine and in their sensitivity to methyl methanesulfonate and to incorporated [3H]thymidine.
Cancer Res. 39:312-320(1979)
PubMed=464569; DOI=10.1128/AAC.15.3.420; PMCID=PMC352676
Klein F., Ricketts R.T., Jones W.I., DeArmon I.A., Temple M.J., Zoon K.C., Bridgen P.J.
Large-scale production and concentration of human lymphoid interferon.
Antimicrob. Agents Chemother. 15:420-427(1979)
PubMed=7316467; DOI=10.1111/j.1469-1809.1980.tb00953.x
Povey S., Jeremiah S., Arthur E., Steel M., Klein G.
Differences in genetic stability between human cell lines from patients with and without lymphoreticular malignancy.
Ann. Hum. Genet. 44:119-133(1980)
PubMed=6286763; DOI=10.4049/jimmunol.129.3.1336
Benjamin D., Magrath I.T., Maguire R.T., Janus C., Todd-Kulikowsk H.D., Parsons R.G.
Immunoglobulin secretion by cell lines derived from African and American undifferentiated lymphomas of Burkitt's and non-Burkitt's type.
J. Immunol. 129:1336-1342(1982)
PubMed=6811418; DOI=10.1016/S0171-2985(11)80031-8
Spira G., Koide N., Aman P., Ber R., Klein G.
Truncated mu chain in a Burkitt lymphoma line (P3HR-1) and its fate in various hemapoietic somatic cell hybrids.
Immunobiology 162:199-209(1982)
PubMed=6231253; DOI=10.1002/ijc.2910330407
Ehlin-Henriksson B., Klein G.
Distinction between Burkitt lymphoma subgroups by monoclonal antibodies: relationships between antigen expression and type of chromosomal translocation.
Int. J. Cancer 33:459-463(1984)
PubMed=2580922; DOI=10.1089/jir.1985.5.65
Feinstein S., Traub A., Lazar A., Mizrahi A., Teitz Y.
Studies on cell binding and internalization of human lymphoblastoid (Namalva) interferon.
J. Interferon Res. 5:65-76(1985)
PubMed=2985879; DOI=10.1016/0145-2126(85)90084-0
Drexler H.G., Gaedicke G., Minowada J.
Isoenzyme studies in human leukemia-lymphoma cell lines -- 1 carboxylic esterase.
Leuk. Res. 9:209-229(1985)
PubMed=2995175
Wurm F.M., Polastri G.D., Hilfenhaus J., Harth H., Zankl H.
Long term cultivation of Namalva cells for interferon production: stable cytogenetic markers for identification of cells in spite of drastic chromosomal variation.
Dev. Biol. Stand. 60:393-403(1985)
PubMed=2998993
Steel C.M., Morten J.E.N., Foster E.
The cytogenetics of human B lymphoid malignancy: studies in Burkitt's lymphoma and Epstein-Barr virus-transformed lymphoblastoid cell lines.
IARC Sci. Publ. 60:265-292(1985)
PubMed=3159941; DOI=10.1016/0145-2126(85)90134-1
Drexler H.G., Gaedicke G., Minowada J.
Isoenzyme studies in human leukemia-lymphoma cell lines -- III Beta-hexosaminidase (E.C. 3.2.1.30).
Leuk. Res. 9:549-559(1985)
PubMed=3874327; DOI=10.1016/0145-2126(85)90133-x
Drexler H.G., Gaedicke G., Minowada J.
Isoenzyme studies in human leukemia-lymphoma cells lines -- II. Acid phosphatase.
Leuk. Res. 9:537-548(1985)
PubMed=3997900; DOI=10.1016/S0092-1157(85)80024-x
Whitaker A.M.
The chromosomes of the Namalwa cell line.
J. Biol. Stand. 13:173-175(1985)
PubMed=2415623; DOI=10.4049/jimmunol.136.1.320
Goldmacher V.S., Lambert J.M., Young A.Y., Anderson J., Tinnel N.L., Kornacki M., Ritz J., Blattler W.A.
Expression of the common acute lymphoblastic leukemia antigen (CALLA) on the surface of individual cells of human lymphoblastoid lines.
J. Immunol. 136:320-325(1986)
PubMed=3080238
Sieverts H., Alabaster O., Goldschmidts W., Magrath I.T.
Expression of surface antigens during the cell cycle in different growth phases of American and African Burkitt's lymphoma cell lines.
Cancer Res. 46:1182-1188(1986)
PubMed=3100061; DOI=10.1016/0008-8749(86)90099-7
Benjamin D., Bazar L.S., Wallace B., Jacobson R.J.
Heterogeneity of B-cell growth factor receptor reactivity in healthy donors and in patients with chronic lymphatic leukemia: relationship to B-cell-derived lymphokines.
Cell. Immunol. 103:394-408(1986)
PubMed=3026973; DOI=10.1002/ijc.2910390215
Ehlin-Henriksson B., Manneborg-Sandlund A., Klein G.
Expression of B-cell-specific markers in different Burkitt lymphoma subgroups.
Int. J. Cancer 39:211-218(1987)
PubMed=2830981; DOI=10.1016/0092-8674(88)90530-2
Lawrence J.B., Villnave C.A., Singer R.H.
Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line.
Cell 52:51-61(1988)
PubMed=1915267; DOI=10.1002/j.1460-2075.1991.tb07837.x; PMCID=PMC452998
Farrell P.J., Allan G.J., Shanahan F., Vousden K.H., Crook T.
p53 is frequently mutated in Burkitt's lymphoma cell lines.
EMBO J. 10:2879-2887(1991)
PubMed=2052620; DOI=10.1073/pnas.88.12.5413; PMCID=PMC51883
Gaidano G., Ballerini P., Gong J.Z., Inghirami G., Neri A., Newcomb E.W., Magrath I.T., Knowles D.M., Dalla-Favera R.
p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia.
Proc. Natl. Acad. Sci. U.S.A. 88:5413-5417(1991)
PubMed=1325212; DOI=10.1182/blood.V80.5.1289.1289
Benjamin D., Knobloch T.J., Dayton M.A.
Human B-cell interleukin-10: B-cell lines derived from patients with acquired immunodeficiency syndrome and Burkitt's lymphoma constitutively secrete large quantities of interleukin-10.
Blood 80:1289-1298(1992)
PubMed=1339415; DOI=10.1002/ijc.2910520320
Middleton P.G., Miller S., Ross J.A., Steel C.M., Guy K.
Insertion of SMRV-H viral DNA at the c-myc gene locus of a BL cell line and presence in established cell lines.
Int. J. Cancer 52:451-454(1992)
PubMed=8344493; DOI=10.1096/fasebj.7.10.8344493
Bhatia K.G., Goldschmidts W., Gutierrez M.I., Gaidano G., Dalla-Favera R., Magrath I.T.
Hemi- or homozygosity: a requirement for some but not other p53 mutant proteins to accumulate and exert a pathogenetic effect.
FASEB J. 7:951-956(1993)
PubMed=8402660
O'Connor P.M., Jackman J., Jondle D., Bhatia K.G., Magrath I.T., Kohn K.W.
Role of the p53 tumor suppressor gene in cell cycle arrest and radiosensitivity of Burkitt's lymphoma cell lines.
Cancer Res. 53:4776-4780(1993)
PubMed=8515068; DOI=10.4049/jimmunol.150.12.5418
Jain V.K., Judde J.-G., Max E.E., Magrath I.T.
Variable IgH chain enhancer activity in Burkitt's lymphomas suggests an additional, direct mechanism of c-myc deregulation.
J. Immunol. 150:5418-5428(1993)
PubMed=8176200; DOI=10.4049/jimmunol.152.10.4749
Benjamin D., Sharma V., Knobloch T.J., Armitage R.J., Dayton M.A., Goodwin R.G.
B cell IL-7. Human B cell lines constitutively secrete IL-7 and express IL-7 receptors.
J. Immunol. 152:4749-4757(1994)
PubMed=7849311; DOI=10.1182/blood.V85.4.893.bloodjournal854893
Stranks G., Height S.E., Mitchell P., Jadayel D.M., Yuille M.A.R., De Lord C.F.M., Clutterbuck R.D., Treleaven J.G., Powles R.L., Nacheva E., Oscier D.G., Karpas A., Lenoir G.M., Smith S.D., Millar J.L., Catovsky D., Dyer M.J.S.
Deletions and rearrangement of CDKN2 in lymphoid malignancy.
Blood 85:893-901(1995)
DOI=10.11418/jtca1981.15.4_211
Matsuo Y., Okochi A., Ariyasu T., Iimura E., Ohno T.
Identification of cell lines with variable numbers of tandem repeat (VNTR) amplified by polymerase chain reaction.
Tissue Cult. Res. Commun. 15:211-219(1996)
PubMed=8558920
Dirks W.G., Zaborski M., Jager K., Challier C., Shiota M., Quentmeier H., Drexler H.G.
The (2;5)(p23;q35) translocation in cell lines derived from malignant lymphomas: absence of t(2;5) in Hodgkin-analogous cell lines.
Leukemia 10:142-149(1996)
PubMed=8568269; DOI=10.4049/jimmunol.156.4.1626
Benjamin D., Sharma V., Kubin M., Klein J.L., Sartori A., Holliday J., Trinchieri G.
IL-12 expression in AIDS-related lymphoma B cell lines.
J. Immunol. 156:1626-1637(1996)
PubMed=9192833
Cherney B.W., Bhatia K.G., Sgadari C., Gutierrez M.I., Mostowski H.S., Pike S.E., Gupta G., Magrath I.T., Tosato G.
Role of the p53 tumor suppressor gene in the tumorigenicity of Burkitt's lymphoma cells.
Cancer Res. 57:2508-2515(1997)
PubMed=9473234; DOI=10.1182/blood.V91.5.1680
Klangby U., Okan I., Magnusson K.P., Wendland M., Lind P., Wiman K.G.
p16/INK4a and p15/INK4b gene methylation and absence of p16/INK4a mRNA and protein expression in Burkitt's lymphoma.
Blood 91:1680-1687(1998)
PubMed=9973220
Gutierrez M.I., Cherney B.W., Hussain A., Mostowski H.S., Tosato G., Magrath I.T., Bhatia K.G.
Bax is frequently compromised in Burkitt's lymphomas with irreversible resistance to Fas-induced apoptosis.
Cancer Res. 59:696-703(1999)
PubMed=10739008; DOI=10.1016/S0145-2126(99)00182-4
Inoue K., Kohno T., Takakura S., Hayashi Y., Mizoguchi H., Yokota J.
Frequent microsatellite instability and BAX mutations in T cell acute lymphoblastic leukemia cell lines.
Leuk. Res. 24:255-262(2000)
PubMed=11226526; DOI=10.1016/S0145-2126(00)00121-1
Inoue K., Kohno T., Takakura S., Hayashi Y., Mizoguchi H., Yokota J.
Corrigendum to: Frequent microsatellite instability and BAX mutations in T cell acute lymphoblastic leukemia cell lines Leukemia Research 24 (2000), 255-262.
Leuk. Res. 25:275-278(2001)
PubMed=12145705; DOI=10.1038/sj.leu.2402519
Langerak A.W., Moreau E.J., van Gastel-Mol E.J., van der Burg M., van Dongen J.J.M.
Detection of clonal EBV episomes in lymphoproliferations as a diagnostic tool.
Leukemia 16:1572-1573(2002)
PubMed=16960149; DOI=10.1182/blood-2006-06-026500
Mestre-Escorihuela C., Rubio-Moscardo F., Richter J.A., Siebert R., Climent J., Fresquet V., Beltran E., Agirre X., Marugan I., Marin M., Rosenwald A., Sugimoto K.-j., Wheat L.M., Karran E.L., Garcia J.F., Sanchez-Verde L., Prosper F., Staudt L.M., Pinkel D., Dyer M.J.S., Martinez-Climent J.A.
Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas.
Blood 109:271-280(2007)
PubMed=18357372; DOI=10.3892/or.19.4.889
Pop I., Pop L., Vitetta E.S., Ghetie M.-A.
Generation of multidrug resistant lymphoma cell lines stably expressing P-glycoprotein.
Oncol. Rep. 19:889-895(2008)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20454443; DOI=10.1155/2010/904767; PMCID=PMC2861168
Uphoff C.C., Denkmann S.A., Steube K.G., Drexler H.G.
Detection of EBV, HBV, HCV, HIV-1, HTLV-I and -II, and SMRV in human and other primate cell lines.
J. Biomed. Biotechnol. 2010:904767.1-904767.23(2010)
PubMed=21269460; DOI=10.1186/1752-0509-5-17; PMCID=PMC3039570
Burkard T.R., Planyavsky M., Kaupe I., Breitwieser F.P., Burckstummer T., Bennett K.L., Superti-Furga G., Colinge J.
Initial characterization of the human central proteome.
BMC Syst. Biol. 5:17.1-17.13(2011)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22885699; DOI=10.1038/nature11378; PMCID=PMC3609867
Schmitz R., Young R.M., Ceribelli M., Jhavar S., Xiao W.-M., Zhang M.-L., Wright G., Shaffer A.L. 3rd, Hodson D.J., Buras E., Liu X.-L., Powell J.I., Yang Y.-D., Xu W.-H., Zhao H., Kohlhammer H., Rosenwald A., Kluin P.M., Muller-Hermelink H.-K., Ott G., Gascoyne R.D., Connors J.M., Rimsza L.M., Campo E., Jaffe E.S., Delabie J., Smeland E.B., Ogwang M.D., Reynolds S.J., Fisher R.I., Braziel R.M., Tubbs R.R., Cook J.R., Weisenburger D.D., Chan W.-C., Pittaluga S., Wilson W., Waldmann T.A., Rowe M., Mbulaiteye S.M., Rickinson A.B., Staudt L.M.
Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics.
Nature 490:116-120(2012)
PubMed=24590883; DOI=10.1002/gcc.22161
Murga Penas E.-M., Schilling G., Behrmann P., Klokow M., Vettorazzi E., Bokemeyer C., Dierlamm J.
Comprehensive cytogenetic and molecular cytogenetic analysis of 44 Burkitt lymphoma cell lines: secondary chromosomal changes characterization, karyotypic evolution, and comparison with primary samples.
Genes Chromosomes Cancer 53:497-515(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893; PMCID=PMC4355981
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25355872; DOI=10.1128/JVI.02570-14; PMCID=PMC4301145
Cao S.-B., Strong M.J., Wang X., Moss W.N., Concha M., Lin Z., O'Grady T., Baddoo M., Fewell C., Renne R., Flemington E.K.
High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the Cancer Cell Line Encyclopedia project.
J. Virol. 89:713-729(2015)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=29892436; DOI=10.1098/rsos.172472; PMCID=PMC5990783
Shioda S., Kasai F., Watanabe K., Kawakami K., Ohtani A., Iemura M., Ozawa M., Arakawa A., Hirayama N., Kawaguchi E., Tano T., Miyata S., Satoh M., Shimizu N., Kohara A.
Screening for 15 pathogenic viruses in human cell lines registered at the JCRB Cell Bank: characterization of in vitro human cells by viral infection.
R. Soc. Open Sci. 5:172472-172472(2018)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5; PMCID=PMC6167786
Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H.H., Koeffler H.P.
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.
BMC Cancer 18:940.1-940.13(2018)
PubMed=30629668; DOI=10.1371/journal.pone.0210404; PMCID=PMC6328144
Uphoff C.C., Pommerenke C., Denkmann S.A., Drexler H.G.
Screening human cell lines for viral infections applying RNA-Seq data analysis.
PLoS ONE 14:E0210404-E0210404(2019)"