Autophagy
Autophagy is an intracellular degradation pathway. It is the intracellular process of transporting the damaged, degenerated or aged protein to lysosomes for digestion and degradation. During the biological evolution, autophagy is a conservative process and can be found from yeast to plant cells as well as mammal cells with many of these regulatory factors having their homologue proteins found in a number of species. The cell autophagy includes three ways, namely macroautophagy, microautophagy and chaperone-mediated autophagy. Macroautophagy refers to the process that the cytoplasm substances are transported into the lysosomes through forming vesicles. This is used for degradation of the aging or damaged intracellular organelles and proteins.
icroautophagy is the process of direct annexation of intracellular material degradation by the lysosomes. The chaperone-mediated cell autophagy refers to that the soluble intracellular proteins directly enter into the lysosomes to be degraded with the help of the molecular chaperone. Under normal physiological conditions, autophagy can ensure the maintenance of the intracellular homeostasis; upon stress conditions, autophagy can effectively prevent the accumulation of toxic or carcinogenic damaged proteins and organelles, further inhibiting the carcinogenesis of cells (Reggiori F, 2002; Shintani, T, 2004).
Autophagy needs to go through about four stages:
Phase 1: the initiation of phagophore
After the cells receive signals of autophagy induction, there will form a membrane structure somewhere in the cytoplasm and then keep expanding. The structure is initially only in flat edge form, liking an open pocket comprised of a bilayer lipid composition and can be observed under the electron microscope, being called autophagy membrane. This is also called as isolation membrane. It is one of the symbols for the occurrence of the autophagy.
Phase 2: the formation of autophagosome
The autophagy membrane keeps elongating and put all the parts of the cytoplasm which required degradation, including the organelles into the "pocket", and then "close" (closure), becoming a closed spherical autophagosome. The autophagosome is like a large "liposome." Autophagosome can be observed under the electron microscope and is one of the symbols of the occurrence of the autophagy. There are two features of autophagosome; one is bilayer lipid membrane; the other is containing cytoplasmic components such as mitochondria and endoplasmic reticulum debris.
Phase 3: transport and fusion of the autophagosome.
At this stage, the formed autophagosome can be infused with the intracellular phagocytic vacuole, phagocytic vesicle and endosome in charge of endocytosis, forming the amphisome with the fusion of lysosome and endosome. But these conditions are not indispensable during the autophagy process.
Stage 4: The degradation of autophagosome.
Autophagosome is fused with the lysosomes to form autolysosome. During this process, the inner membrane of the autophagosome is degraded by the lysosome enzyme, causing the merging of the content of them two. The inclusion in the autophagosome is also broken down into raw materials such as amino acids and fatty acids. These raw materials are transported into the cytoplasm for being reused by cells while those residues which are not recyclable could be discharged outside the cell or retained in the cytoplasm (Gozuacik D, 2004).
TOR itself is a kind of serine/threonine kinase participating in regulation of the cell cycle, growth and proliferation. Under normal circumstances, TOR, through inhibiting the activity of the autophagy initiating molecules Atg1, achieves its control on autophagy. The homologue protein of TOR in mammalian, the mTOR, upon its activated state, inhibits the function of the autophagy initiating molecule, ULK1 through phosphorylation. TOR/mTOR can form TORC1 / mTORC1 and TORC2 / mTORC2 two complexes. mTORC1 includes mTOR, mLST8, PRAS40 and Raptor. . Raptor is a sensitive component to the drug rapamycin, and thus rapamycin is often used to study autophagy for specific inhibition of mTORC1 activity and induction of autophagy. Furthermore, mTORC1, through bio-4E-BP1 and S6K1, can regulate protein synthesis and the biogenesis of ribosome to further regulate cell growth and proliferation. mTORC2 includes mTOR, mSin1 and Rictor is rapamycin-insensitive. mTORC2, through its phosphorylation of Akt (protein kinase B) and PKC (protein kinase C), transfer the signal to the small GTP enzyme Rac1 and RhoA, participating into regulating the formation of the cytoskeleton. AMPK is a protein kinase involved in sensing the intracellular energy state of cell for regulating the metabolism. It also plays an important role in the regulation of autophagy. Upon the low ATP level status (such as hunger or hypoxia) AMPK can sense the change in the level of AMP to be activated, thereby enhancing the inhibitory effect of TSC1/2 on the RheB through phosphorylation, and finally inhibiting the mTOR activity and inducing the autophagy process. In addition, there are also studies which have shown that, AMPK can directly phosphorylate Raptor and inhibit its activity, resulting in a decreased activity of mTORC1. TSC1/ 2 can also integrate the signals from PI3K-AKT and Raf-1-MEK1 / 2-ERK1 / 2, and transmit it to mTORC1. When being subjected to stimuli such as growth factors, Akt is activated, thereby resulting in phosphorylation of TSC2 and inhibition of its binding with TSC1, Raf-1-MEK1 / 2-ERK1/2 can also inhibit the TSC1/TSC2, ultimately activating the Rheb-mTORC1, exerting an inhibitory effect on the cell autophagy process (Meijer AJ, 2004; Liang XH, 1999; Paglin S, 2001).
- Structure:
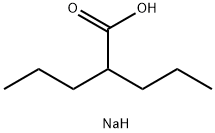
- Chemical Name:Divalproex sodium
- CAS:76584-70-8
- MF:C8H17NaO2
- Structure:
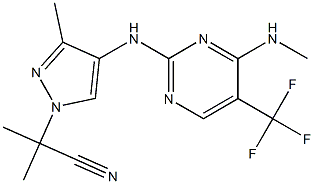
- Chemical Name:GNE-0877
- CAS:1374828-69-9
- MF:C14H16F3N7
- Structure:
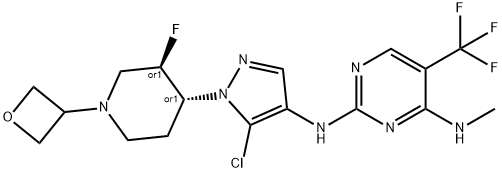
- Chemical Name:GNE-9605
- CAS:1536200-31-3
- MF:C17H20ClF4N7O
- Structure:
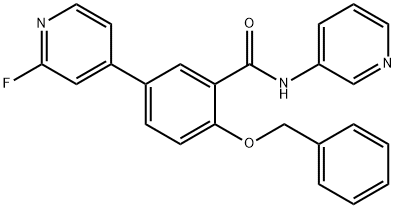
- Chemical Name:LRRK2-kinase inhibitor
- CAS:1285515-21-0
- MF:C24H18FN3O2
- Structure:
- Chemical Name:GNE 7915
- CAS:1351761-44-8
- MF:C19H21F4N5O3