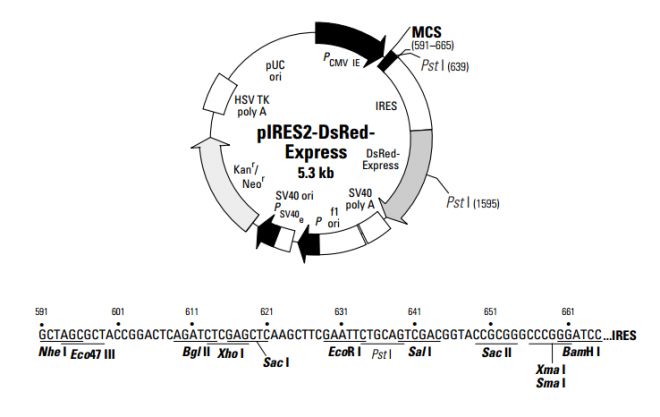
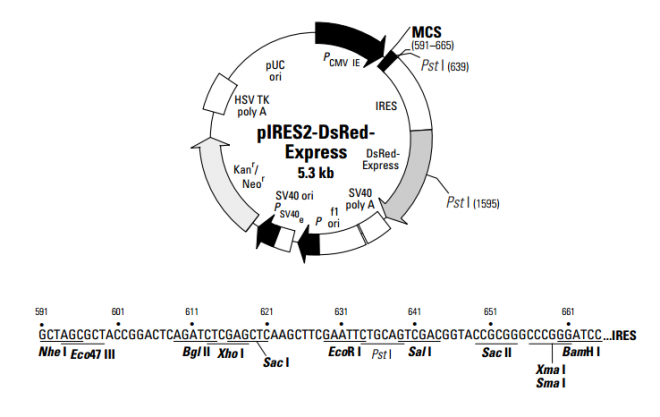
pIRES2-DsRed-Express 载体
pIRES2-DsRed-Express
¥1280
2ug
起订
上海 更新日期:2026-03-06
产品详情:
- 中文名称:
- pIRES2-DsRed-Express 载体
- 英文名称:
- pIRES2-DsRed-Express
- 保存条件:
- 常温运输
- 纯度规格:
- 99%
- 产品类别:
- 质粒/载体
公司简介
上海泽叶生物科技有限公司是一家服务于生命科学领域的高新技术企业,主要为科研机构、高校、院所及企业产品开发研究提供所需要的科研类试剂、耗材、仪器、技术服务等。包括分子生物学、免疫学、微生物学、细胞学、材料科学,通用试剂、药物合成试剂、手性化合物、催化剂及配体、分析试剂、生物试剂,检测试剂等等
| 成立日期 | (10年) |
| 注册资本 | 100万元整 |
| 员工人数 | 1-10人 |
| 年营业额 | ¥ 100万以内 |
| 经营模式 | 贸易,试剂,定制,服务 |
| 主营行业 | 生物化工,有机原料,化学试剂,医药原料,技术服务 |
pIRES2-DsRed-Express 载体相关厂家报价
-
- pCMV-N-DsRed (红色荧光蛋白)
- 上海康朗生物科技有限公司 VIP
- 2026-03-06
- ¥888
-

- 镀锌高温载体
- 武汉兴众诚科技有限公司 VIP
- 2026-03-06
- ¥20
-

- 酸性镀锌低泡高温载体/氯化钾镀锌载体OX-105
- 武汉吉和昌新材料股份有限公司 VIP
- 2026-03-06
- ¥15