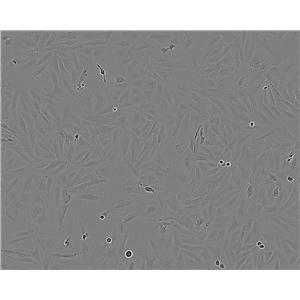
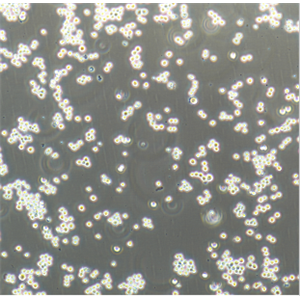
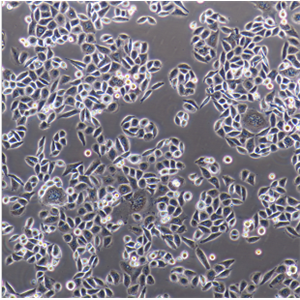
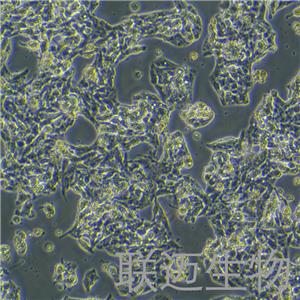
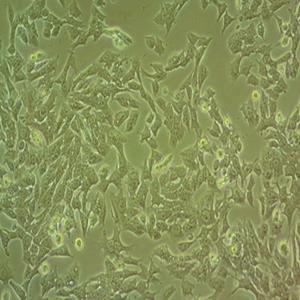
"Hep G2人肝癌复苏细胞保种中心|带STR证书
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
细胞复苏相关注意事项:1.取细胞的过程中注意带HAO防冻手套,护目镜。此项尤为重要,细胞冻存管可能漏入,解冻时冻存管中的气温急剧上升,可导致爆炸。2.冻存的问题:冻存的配置已是常识,在这里不作详述,但二甲基亚砜(DMSO )对细胞不是完全无毒副作用,在常温下,二甲基亚砜对细胞的毒副作较大,因此,必须在1-2min内使冻存完全融化。如果复苏温度太慢,会造成细胞的损伤,二甲基亚砜(DMSO)ZuiHAO选择进口产品。3.离心前须加入少量培养。细胞解冻后二甲基亚砜浓度较GAO,注意加入少量培养可稀释其浓度,以减少对细胞的损伤。4.离心问题:目前主要有两种见解。一种是解冻后的细胞悬直接吹打均匀后分装到培养瓶中进行培养,第二天换。因为离心的目的是两个,去除DMSO,去除死细胞,这个是标准流程,但对一般人来说,把握不HAO离心转速和时间,转的不够活细胞沉底的少,细胞就全被扔掉了,转过了活细胞会受压过大,死亡。此外在操作过程中容易污染,所以不推荐。另一种说法为细胞悬中含有二甲基亚砜(DMSO),DMSO对细胞有一定的毒副作用,所以须将离心后的体前倒净,且一定倒干净。我在试验中按照常规的离心分装的方法进行复苏,结果无异常。5.细胞贴壁少的问题:教科书中说明冻存细胞解冻时1ml细胞要加10ml-15ml培养,而在我的试验中的经验总结为培养基越少细胞越容易贴附。6.复苏细胞分装的问题:试验中我的经验总结为复苏1管细胞一般可分装到1-2只培养瓶中,分装过多,细胞浓度过低,不利于细胞的贴壁。7.加培养基的量放入问题:这个量的多少的把握主要涉及到的问题DMSO的浓度,从如果你加培养基的太少,那么DMSO的浓度就会比较大,就会影响细胞生长,从以前的资料来看,DMSO的浓度在小于0.5%的时候对一般细胞没有什么影响,还有一个说法是1%。所以如果你的冻存的浓度是10%DMSO的话那么加10ml以上的培养基就恰HAO稀释到了无害浓度。
换液周期:每周2-3次
COV504 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:H341细胞、Rh30细胞、Hs 27细胞
KYSE270 Cells;背景说明:详见相关文献介绍;传代方法:1:5传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:2780CP细胞、P3JHR1细胞、BALB/3T3细胞
HCC-44 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NU-GC-2细胞、BLO11细胞、TH1细胞
Hep G2人肝癌复苏细胞保种中心|带STR证书
背景信息:是一种人肝癌细胞系,是来自15岁男性白人的组织,该患者患有高度分化的肝细胞癌;Hep G2细胞形态为上皮细胞样,模式染色体数为55;Hep G2细胞在免疫抑制小鼠中不致瘤。
【细胞培养经验分享】启蒙老师的重要性:一般进实验室都有师兄师姐带着做,他们就是你做细胞的启蒙老师。他们的操作手法、细节、理论讲解就成了你操作的准则,如营养液、细胞瓶的摆放位置、灭菌处理程序、开盖手法、细胞吹打手法等等。要学会他们的正确操作,在第一次的时候就要重视。像养孩子一样养细胞,细胞有时真的很脆弱,最好每天都去看看它,以防止出现培养箱缺水、缺二氧化碳、停电、温度不够等异常现象,也好及时解决这些意外,避免重复实验带来的更大痛苦。好细胞要及时保种:细胞要分批传代,这样即使有一批出了问题,还有一批备用的。像后者一般人可能不容易做到。但这是我血的教训,有一次细胞污染了,全军覆没。当时可后悔没有保种。细胞跟人一样,不同的细胞,培养特性是不一样的。培养过程中要细细体会,不同细胞系使用不同的培养基和血清。
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
OSKM-1 Cells;背景说明:诱导型多能干 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CMEC/D3细胞、Hs 839.T细胞、CCD-841-CoTr细胞
SJSA1 Cells;背景说明:详见相关文献介绍;传代方法:1:5-1:10传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:L2细胞、CL34细胞、SHG44细胞
ME 180 Cells;背景说明:这株细胞来源于一个细胞集落不规则没有显著角质化的侵染性鳞状细胞癌。 单层培养的细胞间可以观察到带状连接,也注意到有细胞质张力丝。 1970年发现支原体污染并去除。 肿瘤坏死因子(TNF)α抑制ME-180的生长。 这株细胞含有人乳头瘤病毒(HPV)DNA,与HPV-39的同源性高于HPV-18。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:RCM-1 [Human rectum adenocarcinoma]细胞、RD 2细胞、IR 983F细胞
Molm 13 Cells;背景说明:急性髓系白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NTERA2-cloneD1细胞、PNT1/A细胞、SK-NSH细胞
Hep G2人肝癌复苏细胞保种中心|带STR证书
物种来源:人源、鼠源等其它物种来源
形态特性:上皮细胞样
有关实验室细胞培养基知识普及:经典的培养基有很多种,Corning、Invirogen(GIBCO)、hermo Fisher(HyClone)等公司都可以提供。其中DMEM、RPMI 1640、MEM、DMEM/F12都是应用Zui广泛的培养基。其他如M199、IMDM、L15培养基等也用于某些细胞的培养;MEM是由Eagle’s基础培养基(BME)发展而来的,其中增加了组分的范围及浓度;BEM(DMEM)培养基是为小鼠成纤维细胞设计的,现在常用于贴壁细胞的培养。DMEM的基浓度是MEM的两倍,维生素浓度是MEM的4倍,采用双倍的HCO3-和CO2浓度起到更HAO的缓冲作用。Zui初的配方中葡萄糖含量为1000mg/L,后来为了某些细胞的生长需要,将葡萄糖含量又调整为4500mg/L,这就是大家常说的低糖和GAO糖了;aMEM含有附加的基、维生素以及核苷和脂肪,它可广泛应用于各种细胞类型,包括对营养成分要求苛刻的细胞;Ham’s F12是为在低血清浓度下克隆CHO细胞而设计的,现在也广泛应用于克隆形成率的分析及原代培养。F12还可以与DMEM等体积混合使用,得到一种GAO浓度与成分多样化相结合的产物,这种培养基已应用于许多原代培养及更难养的细胞系的培养;RPMI 1640培养基是专为淋巴细胞培养而设计的,现在已广泛应用于悬浮细胞的培养。
NMCG1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:HBVSMC细胞、SaOS-2细胞、HEK-EBNA细胞
3T3-J2 Cells;背景说明:胚胎;成纤维 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Potorous tridactylus Kidney 1细胞、C-127细胞、VK2细胞
SCC090 Cells;背景说明:舌鳞癌细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:A673细胞、MOLM13细胞、Lewis Lung细胞
NK-92 MI Cells;背景说明:NK细胞;淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:H2291细胞、GP293细胞、BE(2)M17细胞
HO1-N-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:143TK-细胞、NCIH847细胞、YD15细胞
ACHN Cells;背景说明:该细胞1979年建系,源自一名22岁患有肾细胞腺癌的白人男性的胸腔积液。干扰素可抑制该细胞的生长,该细胞多用于干扰素及其诱导剂的抗增殖研究。;传代方法:1:2-1:3传代,每周2-3次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:HCV 29细胞、BrCL18细胞、MDA-MB-468细胞
MCF-10A Cells;背景说明:乳腺;上皮细胞;自发永生;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NALM6细胞、Human Embryo Kidney-2细胞、MCF7-CTRL细胞
Ramos (RA) Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:BALB/3T3 clone A31细胞、H19-7细胞、Nb-2细胞
ECV304 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H82细胞、3T3L1细胞、GOS-3细胞
HS 445T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MDA.MB.435细胞、PK15细胞、L-cells细胞
NIT-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,每周换液1-2次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:NPA 87细胞、Co-115细胞、HCC202细胞
COLO-829 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:A-10细胞、SW48细胞、CA 46细胞
LAN1 Cells;背景说明:神经母细胞瘤;骨髓转移;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HCE细胞、SKG-IIIa细胞、Mac-1细胞
H-1623 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2-3次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:HOP-92细胞、Caki-2细胞、MFM-223细胞
M109 Cells;背景说明:肺癌;BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:JAR细胞、RGC5细胞、Hs895.T细胞
UM-UC-14 Cells;背景说明:肾癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CF-1 MEF细胞、GL261细胞、ZYM-DIEC02细胞
PC 61.5.3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:alpha TC1 clone 6细胞、SK-MEL-MeWo细胞、U-251-MG细胞
Hep G2人肝癌复苏细胞保种中心|带STR证书
1835_iPS1 Cells(提供STR鉴定图谱)
Abcam HeLa LAMP2 KO Cells(提供STR鉴定图谱)
AG23041 Cells(提供STR鉴定图谱)
BayGenomics ES cell line RRF521 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XG740 Cells(提供STR鉴定图谱)
C0806 Cells(提供STR鉴定图谱)
CW60403 Cells(提供STR鉴定图谱)
Een69 Cells(提供STR鉴定图谱)
GM05521 Cells(提供STR鉴定图谱)
SUNE1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:GM06141B细胞、NCIH1930细胞、NCIH446细胞
NCI-H920 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HLEC-B3细胞、Gingival carcinoma Neck Metastasis细胞、CORL279细胞
SU-DHL-6 Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:6传代,3—4天换液1次;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:SL-1细胞、H2227细胞、B-CPAP细胞
OCI AML5 Cells;背景说明:急性髓系白血病细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:H-1693细胞、Hs-688A-T细胞、X63Ag8.653细胞
Rat-2 Cells;背景说明:成纤维细胞;自发永生;Fischer 344;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:KMS18细胞、Kit225-K6细胞、526细胞
N-9 Cells;背景说明:小胶质细胞;雄性;CD-1;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:半贴壁;形态特性:详见产品说明书;相关产品有:PK15细胞、MSF细胞、CG4细胞
A2215 Cells(提供STR鉴定图谱)
Fu97 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KRC/Y细胞、T 84细胞、LN 229细胞
MOLM-14 Cells;背景说明:急性髓系白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:C-643细胞、OCI-Ly07细胞、PANC0203细胞
Biologics Standards-Cercopithecus-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H-810细胞、KAT-5细胞、MB39细胞
Ramos Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:CAL51细胞、PC-9细胞、C32 mel细胞
H1793 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代 ;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:TGW-nu-1细胞、SVEC 4-10细胞、SK Mel 1细胞
OCILY19 Cells;背景说明:弥漫大B淋巴瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:hFOB 1.19细胞、NCIH2029细胞、Walker-256细胞
GM00215A Cells;背景说明:肺;自发永生;雄性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HOSEpiC细胞、H2122细胞、GM02219D细胞
Oka-C1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H-719细胞、NCI-H322细胞、HHFK细胞
GM22625 Cells(提供STR鉴定图谱)
HAP1 MGME1 (-) 2 Cells(提供STR鉴定图谱)
NCI-H2196 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;每周换液2次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MA-104细胞、6-10B细胞、H-1915细胞
J 111 Cells;背景说明:单核细胞白血病;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCI-H2030细胞、MRMT-1细胞、L929细胞
CAL 148 Cells;背景说明:乳腺癌;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:TPC-1细胞、SNK-1细胞、HRA-19细胞
EFM-192A Cells;背景说明:乳腺癌;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H524细胞、Anip 973细胞、Mv1Lu细胞
NS653 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H1869细胞、LAPC-4细胞、PLA-802细胞
NTERA2D1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MARC145细胞、MGH-U3细胞、H2195细胞
HUT125 Cells;背景说明:腺鳞状肺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MLE 12细胞、P3X63Ag8653细胞、32D clone3细胞
HMC Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:CEM-CCRF (CAMR)细胞、624 MEL细胞、GM2132细胞
HPSI1116i-uacs_3 Cells(提供STR鉴定图谱)
KCC-MS871 Cells(提供STR鉴定图谱)
MGC-803/TA Cells(提供STR鉴定图谱)
NIRRHe001-A Cells(提供STR鉴定图谱)
RAW 264.7-EGFP Cells(提供STR鉴定图谱)
Ubigene HEK293 ZFP36 KO Cells(提供STR鉴定图谱)
WON,PY Cells(提供STR鉴定图谱)
HCT116-SLC27A2-KO-c10 Cells(提供STR鉴定图谱)
T2 (174 x CEM.T2) Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:6传代,每周换液2—3次;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:Bowes melanoma cells细胞、NCIH2347细胞、GP2d细胞
hRMECs Cells;背景说明:视网膜微血管;内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HSAS4细胞、LSECs细胞、NCI-H1672细胞
KLM1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:EU-4细胞、ESC-410细胞、NCIH2110细胞
Th17 Cells;背景说明:辅助性T Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:MIN-6细胞、SHIN3细胞、Eph4 1424细胞
NOZAWA Cells;背景说明:患者有癌性腹膜炎。细胞为中等分化的管状胆囊癌。会分泌AFP和CEA。倍增时间48小时,板植率14-19%。细胞可在裸鼠中成瘤,形态与原发肿瘤相似。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:上皮细胞样;相关产品有:ME180细胞、NCI-SNU-878细胞、HepaRG细胞
NOZAWA Cells;背景说明:患者有癌性腹膜炎。细胞为中等分化的管状胆囊癌。会分泌AFP和CEA。倍增时间48小时,板植率14-19%。细胞可在裸鼠中成瘤,形态与原发肿瘤相似。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:上皮细胞样;相关产品有:ME180细胞、NCI-SNU-878细胞、HepaRG细胞
LOU-NH-91 Cells;背景说明:肺鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MCF-7/AdrR细胞、SUP-M2细胞、C-33-A细胞
KYSE-50 Cells;背景说明:食管鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs604T细胞、Calu 3细胞、SW 620细胞
NCI-H250 Cells;背景说明:小细胞肺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:半贴壁;形态特性:详见产品说明书;相关产品有:H-295细胞、H1341细胞、NCI-H226细胞
MGH-U1 (EJ) Cells;背景说明:膀胱癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Lewis lung carcinoma line 1细胞、HBL-100细胞、H1436细胞
NP-69 Cells;背景说明:鼻咽;上皮细胞;SV40转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-4-II-E细胞、Huh7.5细胞、NCIH1105细胞
NRK-49F Cells;背景说明:肾;成纤维细胞;自发永生;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SK-MEL 28细胞、SK-N-BE细胞、MCCAR细胞
Eph4 1424 Cells;背景说明:乳腺癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HDMEC细胞、RBMVEC细胞、LC-2-Ad细胞
HSKMC Cells;背景说明:骨骼肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:C8166-CD4细胞、MD Anderson-Metastatic Breast-157细胞、SIRC细胞
LNCaP-Clone-FGC Cells;背景说明:人前列腺癌细胞LNCaP克隆FGC是从一位50岁白人男性(血型B+)的左锁骨淋巴结针刺活检中分离,该患者经确诊为前列腺癌转移。 这株细胞对5-α-二睾酮(生长调节子和酸性脂酶产物)有响应。这株细胞并不形成一致的单层,而是形成集落,在传代时可以用滴管反复吹吸打碎。它们仅仅轻轻地吸附在基底上,不形成汇合,很快使培养基变酸。生长很慢。传代后48小时内不应扰动。当培养瓶封包后,多数细胞从培养瓶底分离,悬浮在培养基中。收到后,在通常培养单层细胞的条件下培养24到48小时,以合细胞再贴壁。;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:H1954细胞、3T6 Swiss Albino细胞、Tj-905细胞
STAN131i-214C3 Cells(提供STR鉴定图谱)
WM 451-Lu Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HCC2185细胞、H-716细胞、MKN 45细胞
DR2R1610 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:253J细胞、50.B1细胞、Tregs细胞
NRK 52E Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:OVCAR8细胞、Human Fetal Thymocyte-8810细胞、GC-1细胞
OCI AML5 Cells;背景说明:急性髓系白血病细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:H-1693细胞、Hs-688A-T细胞、X63Ag8.653细胞
DAKIKI Clone 1 Cells;背景说明:B淋巴细胞;EBV转染;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:A-431细胞、766T细胞、LLC-MK-2细胞
4175 Cells;背景说明:乳腺癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:OVCAR433细胞、MDCK II细胞、Human Melanoma Cell Bowes细胞
University of Arizona Cell Culture-812 Cells;背景说明:该细胞是由Liebovitz A等于1986年从一名43岁的白人女性乳腺导管癌患者的乳腺切除肿瘤组织中分离建立的;手术前该病人曾接受过广泛的化疗。该细胞HER-2/neu癌基因序列有15倍的扩增;雌激素受体ER、孕激素受体PR和糖蛋白P阴性。;传代方法:1:3传代;5-7天1次。 ;生长特性:贴壁生长;形态特性:上皮样;相关产品有:WEHI 231细胞、HCC-1143细胞、NCIH460细胞
Stanford University-Diffuse Histiocytic Lymphoma-16 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:EFM19细胞、DHL-2细胞、U937细胞
GM-3573 Cells;背景说明:这是P3X63Ag8(ATCCTIB-9)的一个不分泌克隆。Kappa链合成了但不分泌。能抗0.1mM8-氮杂鸟嘌呤但不能在HAT培养基中生长。据报道它是由于缺失了3-酮类固醇还原酶活性的胆固醇营养缺陷型。检测表明肢骨发育畸形病毒(鼠痘)阴性。;传代方法:1:2传代,3天内可长满。;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:CCD18细胞、COLO-394细胞、CCRF-CEM C7细胞
KYSE50 Cells;背景说明:食管鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HMVEC细胞、NCIH548细胞、T173细胞
OCIAML5 Cells;背景说明:急性髓系白血病细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:Leukemia 1210细胞、HIT T15细胞、COLO 320 DM细胞
OS-732 Cells;背景说明:骨肉瘤;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:6T-CEM细胞、no-11细胞、SVOG细胞
NCIH810 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:6传代,每周2-3次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:LS174细胞、A673细胞、TB-1 Lu细胞
BIU-87 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:3T3L1细胞、P3X63 AG8-653细胞、PLA802细胞
Hep G2人肝癌复苏细胞保种中心|带STR证书
BayGenomics ES cell line HMA339 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XB522 Cells(提供STR鉴定图谱)
CL18/1D8 Cells(提供STR鉴定图谱)
MANEM1-5D10 Cells(提供STR鉴定图谱)
S-163-6 Cells(提供STR鉴定图谱)
WB311 Cells(提供STR鉴定图谱)
" "Patent=US4393133
Knowles B.B., Aden D.P.
Human hepatoma derived cell line, process for preparation thereof, and uses therefor.
Patent number US4393133, 12-Jul-1983
PubMed=2439335; DOI=10.1111/j.1432-1033.1987.tb11497.x
Vincent C., Marceau M., Blangarin P., Bouic P., Madjar J.-J., Revillard J.-P.
Purification of alpha 1-microglobulin produced by human hepatoma cell lines. Biochemical characterization and comparison with alpha 1-microglobulin synthesized by human hepatocytes.
Eur. J. Biochem. 165:699-704(1987)
PubMed=8224613; DOI=10.1096/fasebj.7.14.8224613
Puisieux A., Galvin K., Troalen F., Bressac B., Marcais C., Galun E., Ponchel F., Yakicier C., Ji J.-W., Ozturk M.
Retinoblastoma and p53 tumor suppressor genes in human hepatoma cell lines.
FASEB J. 7:1407-1413(1993)
PubMed=8384076; DOI=10.1016/0165-4608(93)90227-D
Chen H.-L., Chiu T.-S., Chen P.-J., Chen D.-S.
Cytogenetic studies on human liver cancer cell lines.
Cancer Genet. Cytogenet. 65:161-166(1993)
PubMed=8389256; DOI=10.1093/carcin/14.5.987
Hsu I.-C., Tokiwa T., Bennett W.P., Metcalf R.A., Welsh J.A., Sun T.-T., Harris C.C.
p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines.
Carcinogenesis 14:987-992(1993)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045; PMCID=PMC45163
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=8050184; DOI=10.1111/j.1365-2249.1994.tb06089.x; PMCID=PMC1534706
Wadee A.A., Paterson A., Coplan K.A., Reddy S.G.
HLA expression in hepatocellular carcinoma cell lines.
Clin. Exp. Immunol. 97:328-333(1994)
PubMed=8835345; DOI=10.1002/(SICI)1096-9071(199602)48:2<133::aid-jmv3>3.0.CO;2-A
Tsuboi S., Nagamori S., Miyazaki M., Mihara K., Fukaya K.-i., Teruya K., Kosaka T., Tsuji T., Namba M.
Persistence of hepatitis C virus RNA in established human hepatocellular carcinoma cell lines.
J. Med. Virol. 48:133-140(1996)
DOI=10.11418/jtca1981.16.3_173
Mihara K., Miyazaki M., Fushimi K., Tsuji T., Inoue Y., Fukaya K.-i., Ohashi R., Namba M.
The p53 gene status and other cellular characteristics of human cell lines maintained in our laboratory.
Tissue Cult. Res. Commun. 16:173-178(1997)
PubMed=9178645; DOI=10.1006/cimm.1997.1108
Nakao M., Sata M., Saitsu H., Yutani S., Kawamoto M., Kojiro M., Itoh K.
CD4+ hepatic cancer-specific cytotoxic T lymphocytes in patients with hepatocellular carcinoma.
Cell. Immunol. 177:176-181(1997)
PubMed=9359923; DOI=10.18926/AMO/30789
Mihara K., Miyazaki M., Kondo T., Fushimi K., Tsuji T., Inoue Y., Fukaya K.-i., Ishioka C., Namba M.
Yeast functional assay of the p53 gene status in human cell lines maintained in our laboratory.
Acta Med. Okayama 51:261-265(1997)
PubMed=11050057; DOI=10.1053/jhep.2000.19349
Wong N., Lai P.B.-S., Pang E., Leung T.W.-T., Lau J.W.-Y., Johnson P.J.
A comprehensive karyotypic study on human hepatocellular carcinoma by spectral karyotyping.
Hepatology 32:1060-1068(2000)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11981770; DOI=10.1053/jhep.2002.32668
Clemens D.L., Forman A., Jerrells T.R., Sorrell M.F., Tuma D.J.
Relationship between acetaldehyde levels and cell survival in ethanol-metabolizing hepatoma cells.
Hepatology 35:1196-1204(2002)
PubMed=12029633; DOI=10.1053/jhep.2002.33683
Yasui K., Arii S., Zhao C., Imoto I., Ueda M., Nagai H., Emi M., Inazawa J.
TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas.
Hepatology 35:1476-1484(2002)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
DOI=10.1385/CP:1:3-4:313
Pang R.T.-K., Poon T.C.-W., Wong N., Lai P.B.-S., Wong N.L.-Y., Chan C.M.-L., Yu J.W.S., Chan A.T.-C., Sung J.J.-Y.
Comparison of protein expression patterns between hepatocellular carcinoma cell lines and a hepatoblastoma cell line.
Clin. Proteomics 1:313-331(2004)
PubMed=14980111
Zhai B.-J., Wu F., Shao Z.-Y., Hu K., Zhao C.-L., Wang Z.-B.
Establishment of human hepatocellular carcinoma multidrug-resistance cell line (HepG2/Adm) and study apoptosis induced by low-frequency pulse ultrasound exposure.
Zhonghua Gan Zang Bing Za Zhi 12:95-98(2004)
PubMed=15767549; DOI=10.1158/1535-7163.MCT-04-0234
Nakatsu N., Yoshida Y., Yamazaki K., Nakamura T., Dan S., Fukui Y., Yamori T.
Chemosensitivity profile of cancer cell lines and identification of genes determining chemosensitivity by an integrated bioinformatical approach using cDNA arrays.
Mol. Cancer Ther. 4:399-412(2005)
PubMed=16181800; DOI=10.1016/j.biocel.2005.07.010
Donohue T.M., Osna N.A., Clemens D.L.
Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity.
Int. J. Biochem. Cell Biol. 38:92-101(2006)
PubMed=16935386; DOI=10.1016/j.jhep.2006.05.019
Sun D.-X., Nassal M.
Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus.
J. Hepatol. 45:636-645(2006)
PubMed=17254797; DOI=10.1016/j.biologicals.2006.10.001
Azari S., Ahmadi N., Jeddi-Tehrani M., Shokri F.
Profiling and authentication of human cell lines using short tandem repeat (STR) loci: report from the National Cell Bank of Iran.
Biologicals 35:195-202(2007)
PubMed=19215227; DOI=10.1111/j.1349-7006.2009.01082.x; PMCID=PMC11158180
Kuwahara Y., Li L., Baba T., Nakagawa H., Shimura T., Yamamoto Y., Ohkubo Y., Fukumoto M.
Clinically relevant radioresistant cells efficiently repair DNA double-strand breaks induced by X-rays.
Cancer Sci. 100:747-752(2009)
PubMed=19751877; DOI=10.1016/j.humpath.2009.07.003
Lopez-Terrada D.H., Cheung S.-W., Finegold M.J., Knowles B.B.
Hep G2 is a hepatoblastoma-derived cell line.
Hum. Pathol. 40:1512-1515(2009)
PubMed=20069059; DOI=10.1155/2010/437143; PMCID=PMC2801507
Srisomsap C., Sawangareetrakul P., Subhasitanont P., Chokchaichamnankit D., Chiablaem K., Bhudhisawasdi V., Wongkham S., Svasti J.
Proteomic studies of cholangiocarcinoma and hepatocellular carcinoma cell secretomes.
J. Biomed. Biotechnol. 2010:437143.1-437143.18(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20228232; DOI=10.1124/dmd.109.031831; PMCID=PMC2879958
Hart S.N., Li Y., Nakamoto K., Subileau E.-A., Steen D., Zhong X.-B.
A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues.
Drug Metab. Dispos. 38:988-994(2010)
PubMed=20937217; DOI=10.1170/149
Di Masi A., Viganotti M., Antoccia A., Magrelli A., Salvatore M., Azzalin G., Tosto F., Lorenzetti S., Maranghi F., Mantovani A., Macino G., Tanzarella C., Taruscio D.
Characterization of HuH6, Hep3B, HepG2 and HLE liver cancer cell lines by WNT/beta-catenin pathway, microRNA expression and protein expression profile.
Cell. Mol. Biol. 56:OL1299-OL1317(2010)
PubMed=21269460; DOI=10.1186/1752-0509-5-17; PMCID=PMC3039570
Burkard T.R., Planyavsky M., Kaupe I., Breitwieser F.P., Burckstummer T., Bennett K.L., Superti-Furga G., Colinge J.
Initial characterization of the human central proteome.
BMC Syst. Biol. 5:17.1-17.13(2011)
PubMed=22278370; DOI=10.1074/mcp.M111.014050; PMCID=PMC3316730
Geiger T., Wehner A., Schaab C., Cox J., Mann M.
Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins.
Mol. Cell. Proteomics 11:M111.014050-M111.014050(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=23325432; DOI=10.1101/gr.147942.112; PMCID=PMC3589544
Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., Absher D.M., Wold B.J., Myers R.M.
Dynamic DNA methylation across diverse human cell lines and tissues.
Genome Res. 23:555-567(2013)
PubMed=23505090; DOI=10.1002/hep.26402
Wang K., Lim H.Y., Shi S., Lee J., Deng S.-B., Xie T., Zhu Z., Wang Y.-L., Pocalyko D., Yang W.J., Rejto P.A., Mao M., Park C.-K., Xu J.-C.
Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma.
Hepatology 58:706-717(2013)
PubMed=23887712; DOI=10.1038/ncomms3218; PMCID=PMC3731665
Nault J.-C., Mallet M., Pilati C., Calderaro J., Bioulac-Sage P., Laurent C., Laurent A., Cherqui D., Balabaud C., Zucman-Rossi J.
High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions.
Nat. Commun. 4:2218.1-2218.7(2013)
PubMed=24116068; DOI=10.1371/journal.pone.0075692; PMCID=PMC3792989
Weiskirchen R., Weimer J., Meurer S.K., Kron A., Seipel B., Vater I., Arnold N., Siebert R., Xu L.-M., Friedman S.L., Bergmann C.
Genetic characteristics of the human hepatic stellate cell line LX-2.
PLoS ONE 8:E75692-E75692(2013)
PubMed=24618588; DOI=10.1371/journal.pone.0091433; PMCID=PMC3950186
Chernobrovkin A.L., Zubarev R.A.
Detection of viral proteins in human cells lines by xeno-proteomics: elimination of the last valid excuse for not testing every cellular proteome dataset for viral proteins.
PLoS ONE 9:E91433-E91433(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893; PMCID=PMC4355981
Boegel S., Lower M., Bukur T., Sahin U., Castle J.C.
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines.
OncoImmunology 3:e954893.1-e954893.12(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25574106; DOI=10.3748/wjg.v21.i1.311; PMCID=PMC4284350
Cevik D., Yildiz G., Ozturk M.
Common telomerase reverse transcriptase promoter mutations in hepatocellular carcinomas from different geographical locations.
World J. Gastroenterol. 21:311-317(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26160117; DOI=10.1093/toxsci/kfv136; PMCID=PMC4583060
Sison-Young R.L.C., Mitsa D., Jenkins R.E., Mottram D., Alexandre E., Richert L., Aerts H., Weaver R.J., Jones R.P., Johann E., Hewitt P.G., Ingelman-Sundberg M., Goldring C.E.P., Kitteringham N.R., Park B.K.
Comparative proteomic characterization of 4 human liver-derived single cell culture models reveals significant variation in the capacity for drug disposition, bioactivation, and detoxication.
Toxicol. Sci. 147:412-424(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26825538; DOI=10.1016/j.jprot.2016.01.016
Wisniewski J.R., Vildhede A., Noren A., Artursson P.
In-depth quantitative analysis and comparison of the human hepatocyte and hepatoma cell line HepG2 proteomes.
J. Proteomics 136:234-247(2016)
PubMed=27027780; DOI=10.1007/s10565-016-9316-2
Wu Y., Geng X.-C., Wang J.-F., Miao Y.-F., Lu Y.-L., Li B.
The HepaRG cell line, a superior in vitro model to L-02, HepG2 and hiHeps cell lines for assessing drug-induced liver injury.
Cell Biol. Toxicol. 32:37-59(2016)
PubMed=27329724; DOI=10.18632/oncotarget.10161; PMCID=PMC5216950
Watari K., Nishitani A., Shibata T., Noda M., Kawahara A., Akiba J., Murakami Y., Yano H., Kuwano M., Ono M.
Phosphorylation of mTOR Ser2481 is a key target limiting the efficacy of rapalogs for treating hepatocellular carcinoma.
Oncotarget 7:47403-47417(2016)
PubMed=27470094; DOI=10.1016/j.chroma.2016.07.042
Liu Z.-Y., Wang F.-J., Chen J., Zhou Y., Zou H.-F.
Modulating the selectivity of affinity absorbents to multi-phosphopeptides by a competitive substitution strategy.
J. Chromatogr. A 1461:35-41(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=29610054; DOI=10.1016/j.dmpk.2018.03.003; PMCID=PMC6309175
Shi J., Wang X.-W., Lyu L.-Y., Jiang H., Zhu H.-J.
Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: Focusing on drug-metabolizing enzymes.
Drug Metab. Pharmacokinet. 33:133-140(2018)
PubMed=29660373; DOI=10.1016/j.bbagen.2018.04.012
Touat-Hamici Z., Bulteau A.-L., Bianga J., Jean-Jacques H., Szpunar J., Lobinski R., Chavatte L.
Selenium-regulated hierarchy of human selenoproteome in cancerous and immortalized cells lines.
Biochim. Biophys. Acta 1862:2493-2505(2018)
PubMed=30629668; DOI=10.1371/journal.pone.0210404; PMCID=PMC6328144
Uphoff C.C., Pommerenke C., Denkmann S.A., Drexler H.G.
Screening human cell lines for viral infections applying RNA-Seq data analysis.
PLoS ONE 14:E0210404-E0210404(2019)
PubMed=30864654; DOI=10.1093/nar/gkz169; PMCID=PMC6486628
Zhou B., Ho S.S., Greer S.U., Spies N., Bell J.M., Zhang X.-L., Zhu X.-W., Arthur J.G., Byeon S., Pattni R., Saha I., Huang Y.-L., Song G., Perrin D., Wong W.H., Ji H.P., Abyzov A., Urban A.E.
Haplotype-resolved and integrated genome analysis of the cancer cell line HepG2.
Nucleic Acids Res. 47:3846-3861(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31063779; DOI=10.1053/j.gastro.2019.05.001
Caruso S., Calatayud A.-L., Pilet J., La Bella T., Rekik S., Imbeaud S., Letouze E., Meunier L., Bayard Q., Rohr-Udilova N., Peneau C., Grasl-Kraupp B., de Koning L., Ouine B., Bioulac-Sage P., Couchy G., Calderaro J., Nault J.-C., Zucman-Rossi J., Rebouissou S.
Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response.
Gastroenterology 157:760-776(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31378681; DOI=10.1016/j.ccell.2019.07.001; PMCID=PMC7505724
Qiu Z.-X., Li H., Zhang Z.-T., Zhu Z.-F., He S., Wang X.-J., Wang P.-C., Qin J.-J., Zhuang L.-P., Wang W., Xie F.-B., Gu Y., Zou K.-K., Li C., Li C., Wang C.-H., Cen J., Chen X.-T., Shu Y.-J., Zhang Z., Sun L.-L., Min L.-H., Fu Y., Huang X.-W., Lv H., Zhou H., Ji Y., Zhang Z.-G., Meng Z.-Q., Shi X.-L., Zhang H.-B., Li Y.-X., Hui L.-J.
A pharmacogenomic landscape in human liver cancers.
Cancer Cell 36:179-193.e11(2019)
PubMed=31395879; DOI=10.1038/s41467-019-11415-2; PMCID=PMC6687785
Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., van 't Veer L.J., Butte A.J., Goldstein T., Sirota M.
Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types.
Nat. Commun. 10:3574.1-3574.11(2019)
PubMed=31561354; DOI=10.3233/CH-199226; PMCID=PMC6918903
Schulz C., Kammerer S., Kupper J.-H.
NADPH-cytochrome P450 reductase expression and enzymatic activity in primary-like human hepatocytes and HepG2 cells for in vitro biotransformation studies.
Clin. Hemorheol. Microcirc. 73:249-260(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023; PMCID=PMC7339254
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)"