Nuclear transcription factor (NF-κB)
Nuclear transcription factor NF-κB is a kind of critical nuclear transcription factor, usually being presented in virtually the cytoplasm of all kinds of cells in the homo- or heterodimers inactive forms. It has a very important function. It is related to the activation of immune cells, the development of T and B lymphocyte, stress response, apoptosis and many other cellular activities. Many factors can be used to activate the nuclear transcription factor NF-κB, so that they can be transferred from the cytoplasm to nucleus, binding to the κB sites of NF-κB responsive genes to participate into the transcriptional regulation of NF-κB responsive genes.
Nuclear transcription factor NF-κB was originally found in B lymphocytes, binding with the B site of the enhancer of the κ light chain immunoglobulin coding gene, further regulating the transcription of the immunoglobulin κ light chain, thus being named as nuclear transcription factor κB. Members in the nuclear transcription factor NF-κB family usually form complexes with its inhibitory protein IκBs in the form of homodimers or heterodimers, existing in the cytoplasm in its inactive form. The NF -κB can be activated only under the effect of various kinds of activating factors. There are various factors that can activate nuclear transcription factor NF-κB, including various stress stimuli, mucopolysaccharides bacteria, viruses, oxygen free radicals and a variety of cytokines. Their mechanisms of activating the nuclear transcription factor NF-κB also vary.
With the rapid development of molecular biology in recent years, the activation mechanisms of the nuclear transcription factor NF-κB has gradually become clear. The subunit of NF-κB subunit p50, p65, p52, c-Rel and RelB can polymerize to form homo- or heterodimers. Being different with other DNA binding proteins, the subunits of NF-κB can form 2 Ig-like domains in its Rel homology region RHD. The major function of the carboxyl termini of NF-κB is polymerizing two subunits with the main role of the amino-termini being forming a sequence-specific pocket-like structure to specially bind to the κB site of the target genes. On the other hand, the ARD (ankyrin repeat domain) of the inhibitory protein IκBs can form a gently curved cylindrical structure. It contains the amino acid residues that can specifically recognize NF-κB, thus binding to the Ig-like domain of the polymerized p50 / p65. There are six kinds of IκBs in total, namely IκBα, -β, -γ, -ε, -δ, bcl-3. Binding of IκBs to the p65 subunit leads to the conformational change of p65. The amino terminus of p65 subunit Ig-like domain rotate 180 ° so that the key target DNA bound amino acid residues are hidden, thus suppressing the specific binding between the NF- κB with the DNA regulatory regions.
The amino-terminal serine residues at position 32 and 36 of IκBα as well as the serine residues in position 19 and 23 of IκBβ can be subject to phosphorylation by the IκB kinase (IKKα-IKKβ). The phosphorylated IκBs is further ubiquitinated and subject to degradation by the proteasome, making the NF-κB be dissociated from the NF-κB-IκBs complexes and be translocated into the nucleus, binding to the corresponding target genes. However, at the same, IκBs can be rapidly re-synthesized after being degraded and can enter the nucleus to bind with NF-κB while the formation of NF-κB-IκBs complex makes NF-κB be dissociated from its binding sites on DNA κB and re-translocated in the cytoplasm, thereby realizing the cycle of NF-κB activation and deactivation, completing the function of NF-κB as a nuclear transcription factors for regulating gene transcription. There are 60 kinds of gene transcription requires the participation of the nuclear transcription factor NF-κB such as genes related to cell adhesion, immune stimulation, apoptosis, inflammatory cell chemotaxis, cell differentiation, extracellular matrix degradation and other related genes. The molecular basis of these important physiological processes is the continuous cycle of NF-κB activation and deactivation.
- Structure:
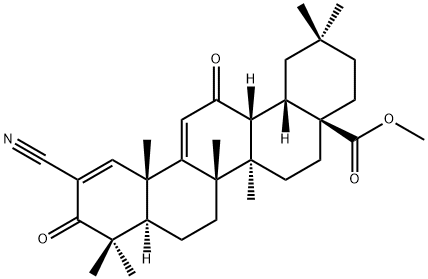
- Chemical Name:Bardoxolone methyl
- CAS:218600-53-4
- MF:C32H43NO4
- Structure:
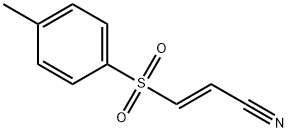
- Chemical Name:BAY 11-7082
- CAS:19542-67-7
- MF:C10H9NO2S
- Structure:
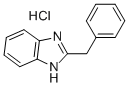
- Chemical Name:2-benzyl-1H-benzimidazole monohydrochloride
- CAS:1212-48-2
- MF:C14H13ClN2
- Chemical Name:ATF4
- CAS:
- MF:
- Structure:
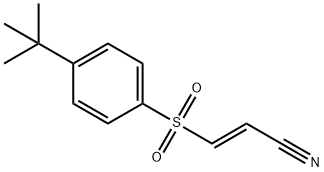
- Chemical Name:BAY 11-7085
- CAS:196309-76-9
- MF:C13H15NO2S
- Structure:
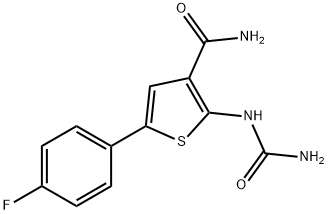
- Chemical Name:TPCA-1
- CAS:507475-17-4
- MF:C12H10FN3O2S
- Structure:
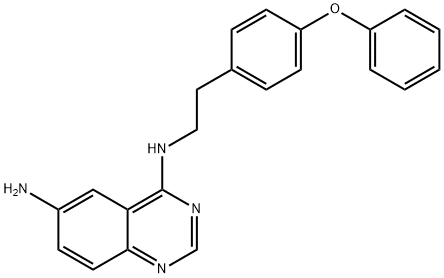
- Chemical Name:6-AMINO-4-(4-PHENOXYPHENYLETHYLAMINO)QUINAZOLINE
- CAS:545380-34-5
- MF:C22H20N4O
- Structure:
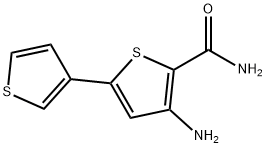
- Chemical Name:SC-514
- CAS:354812-17-2
- MF:C9H8N2OS2
- Structure:

- Chemical Name:4-METHYL-N1-(3-PHENYLPROPYL)BENZENE-1,2-DIAMINE
- CAS:749886-87-1
- MF:C16H20N2
- Structure:
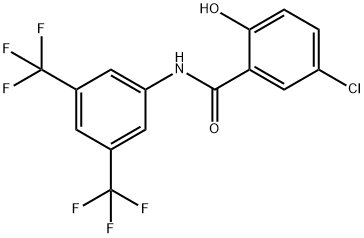
- Chemical Name:IMD-0354
- CAS:978-62-1
- MF:C15H8ClF6NO2
- Structure:
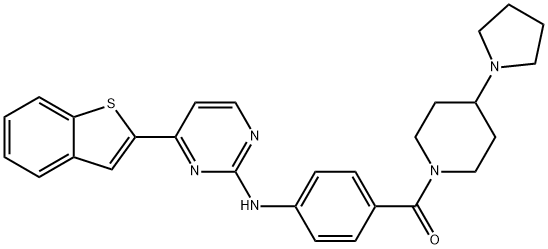
- Chemical Name:IKK 16
- CAS:873225-46-8
- MF:C28H29N5OS
- Structure:
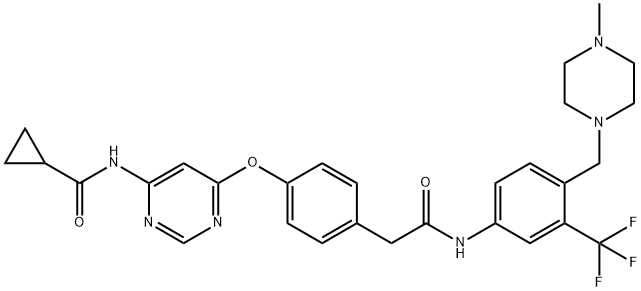
- Chemical Name:WS6
- CAS:1421227-53-3
- MF:C29H31F3N6O3
- Structure:
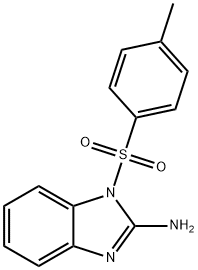
- Chemical Name:Nodinitib-1
- CAS:799264-47-4
- MF:C14H13N3O2S
- Structure:
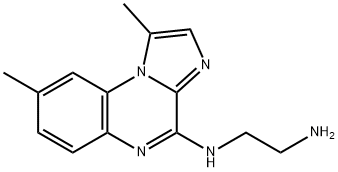
- Chemical Name:IKK Inhibitor III, BMS-345541
- CAS:445430-58-0
- MF:C14H17N5
- Structure:
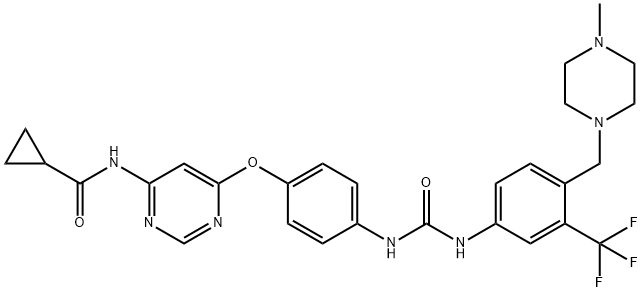
- Chemical Name:WS3
- CAS:1421227-52-2
- MF:C28H30F3N7O3
- Structure:
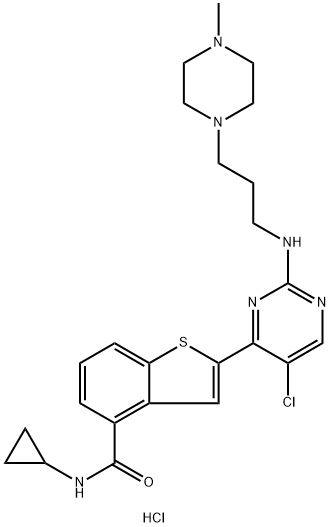
- Chemical Name:LY2409881 trihydrochloride
- CAS:946518-60-1
- MF:C24H30Cl2N6OS
- Structure:
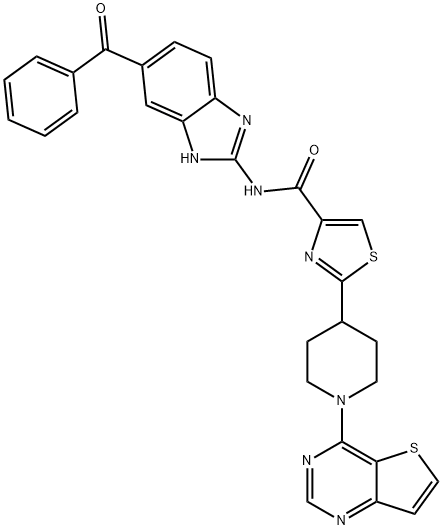
- Chemical Name:4-ThiazolecarboxaMide, N-(6-benzoyl-1H-benziMidazol-2-yl)-2-(1-thieno[3,2-d]pyriMidin-4-yl-4-piperidinyl)-
- CAS:913822-46-5
- MF:C29H23N7O2S2