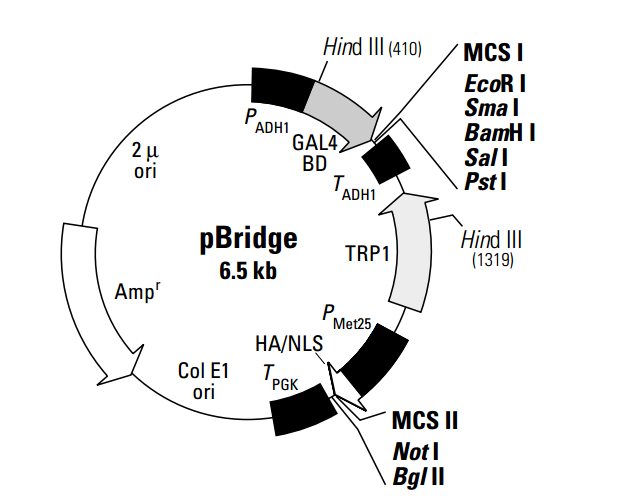
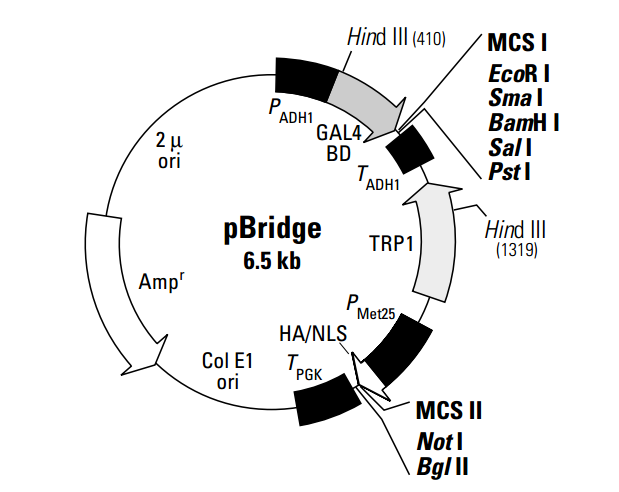
pBridgeTM expresses two proteins: a DNA-binding domain fusion, and an additional protein (1–3). pBridge thus allows establishment of three-hybrid systems when used in combination with an activation domain fusion vector and yeast strains from any of CLONTECH's GAL4-based two-hybrid systems (#K1604-1, #K1605-1, #K1612-1). This vector generates a hybrid protein that contains the sequences for the GAL4 DNA-binding domain (DNA-BD; a.a. 1–147) and the sequence cloned into MCS I. The fusion protein is expressed in yeast host cells from the constitutive ADH1 promoter; transcription is terminated at the ADH1 transcription termination signal. The hybrid protein is targeted to the yeast nucleus by nuclear localization sequences (NLS) that are an intrinsic part of the GAL4 DNA-BD (3). An additional gene of interest can be cloned into MCS II which is located downstream of an HA epitope and a second NLS. The resulting fusion protein is conditionally expressed from the MET25 promoter in response to methionine levels in the medium; i.e., it is repressed in the presence of 1 mM methionine and expressed in the absence of methionine (1).
pBridge is a shuttle vector that replicates autonomously in both E. coli and S. cerevisiae. It carries the bla gene (for ampicillin resistance in E. coli) and the TRP1 nutritional marker that allow yeast auxotrophs carrying pBridge to grow on limiting synthetic medium lacking tryptophan. Note: Yeast strain Y187 is a methionine auxotroph; therefore, haploid Y187 harboring pBridge cannot be grown on medium lacking methionine.
载体应用
The pBridge Vector now makes it possible to investigate ternary protein complex formation (1, 2). pBridge contains two distinct multiple cloning sites to allow expression of the DNA-BD fusion as well as a third protein. When pBridge is used in conjunction with the AD fusion vector and yeast strains from any one of CLONTECH’s GAL4-based two-hybrid systems, the MATCHMAKER Two-Hybrid System (#K1605-1) or the MATCHMAKER Two-Hybrid System 2 & 3 (#K1604-1 & #K1612-1), a ‘three-hybrid’ system can be established that is dependent on the expression of a third protein. The yeast two-hybrid system has proven to be a powerful molecular approach for detecting protein-protein (binary) interactions and has contributed significantly to the dissection of many molecular pathways. CLONTECH’s MATCHMAKER GAL4- and LexA-based two-hybrid systems and cDNA libraries have facilitated the widespread use of this technology. However, the two-hybrid system is designed to detect the interaction between just two proteins, which are expressed as the AD and BD fusions.
The figure below demonstrates the more complex protein interactions that can be investigated with the three-hybrid system.The third protein in this system can participate in the interaction in several ways: as a “bridge”, interacting with two proteins that do not directly interact with each other; to stabilize a weak interaction between two proteins; or as an inhibitor or modifier (e.g., kinase; 3) of one or both of the proteins. Alternatively, a competitor of a two-hybrid interaction can be expressed from this promoter to confirm the specificity of the two-hybrid interaction (1). The conditional expression of the third protein allows investigation of its role in the interaction between the AD and BD fusion proteins. Like the two-hybrid system, the three-hybrid system can be used to screen libraries to clone new interacting partners, either for the third protein or the binding domain fusion.
Expression of the third protein is controlled by a conditional methionine promoter (PMET25) such that it is expressed in the absence of methionine. This allows expression to be switched on or off by a simple replica plating step. The effect of the third protein is indicated by expression of a reporter or nutritional marker. To facilitate experiments with pBridge, we offer three drop-out media supplements that lack methionine: –His/–Leu/–Met/–Trp (#8620-1); –Leu–Met/–Trp (#8621-1); and –Met/–Trp (#8622-1).