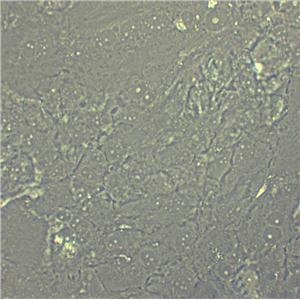
"HT-29人结肠癌复苏细胞保种中心|带STR证书
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
贴壁消化难题:1,先用PBS 把细胞洗两遍,使瓶内没有血清了,减少对胰酶的中和,然后用新配的0.25%的胰酶加入3ml左右,放在37度,然后可以在细胞有些消化下来时,拿着瓶口,运用手腕的力量轻轻震荡瓶内体,这样细胞很快就下来了,还不需要吹打,分散也均匀;2,成团、絮状:消化里加入eda可以减少细胞成团的现象,血清可以终止胰酶的作用,如果是进口血清的话也能终止eda的作用。用胰酶消化后胰酶可以倒掉,也可以不倒,直接加血清终止,如果消化中加入了eda的话,就要将消化倒干净,如果细胞贴壁要求不是很严格的话,一般不需要进行离心。鼻咽癌细胞的贴壁能力很强,用0.5%胰酶(含0.1%EDA)一般要消化12~15min。用PBS洗涤时要洗净残余的培养基,加入胰酶后在培养箱中消化(避免细胞室温下受损以及在此温度时胰酶活性Zui强)至细胞收缩变圆(可显微镜下观察)且有少许细胞脱落(有流下来的趋势),随后立即弃去胰酶(如果脱落的细胞很多且需要大量细胞实验,则不能弃去胰酶),加入培养基仔细吹打(不能用无血清培养基或者PBS替代,否则细胞聚集成团块或絮状)。一般我都离心一次弃去上清(去除残留的胰酶及漂浮的死细胞或细胞碎片);消化过度:马上用培养基中和,用吸管吹打细胞,收集全部的细胞到以无菌的离心管中800RPM 3分钟。弃上清,用全培重悬,换新的培养瓶继续培养,状态不HAO的细胞在培养的过程中会死亡脱落,在换的时候可以清除掉!
换液周期:每周2-3次
KU1919 Cells;背景说明:膀胱癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MC9细胞、NMC-G1细胞、Panc 04.03细胞
MGHU3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MOLT.4细胞、WM-115细胞、NCIH1963细胞
Karpas 299 Cells;背景说明:间变性大细胞淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:HNE-1细胞、AML-12细胞、RINm5F细胞
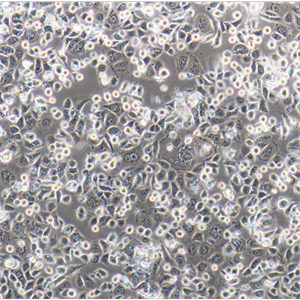
HT-29人结肠癌复苏细胞保种中心|带STR证书
背景信息:该细胞是1964年由FoghJ用移植培养方法和含15%FBS的F12培养从原发性肿瘤分离的。近来,已建株的培养细胞用含血清的McCoy's5a培养基培养。该细胞系在裸鼠中成瘤,也能在类固醇处理的地鼠中成瘤。该细胞可合成IgA、A、TGFβ结合蛋白和黏素;表达尿激酶受体,但没有检测到血浆酶原活性;不表达CD4,但细胞表面表达半乳糖神经酰胺(HIV的可能替代受体)。该细胞系癌基因c-myc、K-ras、H-ras、N-ras、Myb、sis、fos阳性;p53基因过表达,并且在273位密码子处发
公司细胞系形态漂亮、增殖倍数高、纯度高、功能性强,细胞培养就跟养孩子一个样。养孩子要喂奶,养细胞要加补液,都需要在前期补充足够的营养,初始状态的细胞或刚刚复苏的细胞还要适量加入血清或细胞因子来帮助它们的存活增殖,如果营养物质缺乏,细胞就会不生长甚至死亡。养孩子要从小培养学习,养细胞也得培养宝宝顺利生下来,你会经常抚摸他,给他看各种颜色,刺激他的五感。细胞也是一样,分离后的细胞需要使用特定的细胞因子进行活化、增殖。另外加入因子的种类、因子的浓度、加入时间、加入顺序都会影响细胞最终的结果。养孩子最怕孩子生病,养细胞最怕被污染,平时你会仔细观察宝宝是否呕吐、是否突然哭闹,猜测宝宝是否生病了。对于细胞,我们也需要时刻进行观察的,假如培养液浑浊(污染了),则需要换液后加抗生素;假如细胞增殖不明显,形态变差,则可能是因为营养不足了,对贴壁细胞可以消化后重新用新的培养基接种并加倍加入细胞因子含量;对悬浮细胞增殖能力不强的,则不着急补液,只是先补加血清、细胞因子看是否可以好转。培养时还得全程在无菌的环境,一个小小的偏差,细胞就会死亡。
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
WERI Rb-1 Cells;背景说明:WERI-Rb-I细胞株是1974年R.M. McFall 和 T.W. Sery建立的两株人眼癌细胞系中的一株。 细胞能在Difco Bacto-Agar中存活但不形成克隆。 扫描电镜显示在表面囊泡,板状伪足和微绒毛在数量上和频率上的改变。 细胞分化研究,肿瘤治疗的动物模型和生化评价都涉及这株细胞。;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:圆形细胞聚集成葡萄状;相关产品有:LY细胞、SW1116细胞、U-343 MG细胞
RGC6 Cells;背景说明:胶质细胞株C6是由Benda等用N-nitrosomethylurea诱导的大鼠胶质瘤克隆,并经过一系列的体外培养和动物传代交替后建成的。 当细胞从低密度生长到满瓶时,S-100产量增加10倍。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:SF-295细胞、MFE 296细胞、KYSE 180细胞
Large Cell Lung Cancer-103H Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:WPMY-1细胞、A2780CP细胞、NCI-SNU-423细胞
WERI Cells;背景说明:WERI-Rb-I细胞株是1974年R.M. McFall 和 T.W. Sery建立的两株人眼癌细胞系中的一株。 细胞能在Difco Bacto-Agar中存活但不形成克隆。 扫描电镜显示在表面囊泡,板状伪足和微绒毛在数量上和频率上的改变。 细胞分化研究,肿瘤治疗的动物模型和生化评价都涉及这株细胞。;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:圆形细胞聚集成葡萄状;相关产品有:OV3121细胞、AHH1细胞、HAC-84细胞
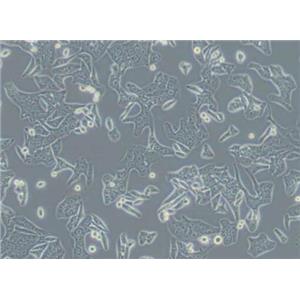
HT-29人结肠癌复苏细胞保种中心|带STR证书
物种来源:人源、鼠源等其它物种来源
形态特性:上皮细胞样
【胰蛋白酶消化操作】1)加入消化:小心吸出旧培养,用PBS清洗(冲洗),加入适量消化(胰蛋白酶),注意消化的量以盖住细胞ZuiHAO,Zui佳消化温度是37℃;2)显微镜下观察细胞:倒置显微镜下观察消化细胞,若胞质回缩,细胞之间不再连接成片,表明此时细胞消化适度;(3)吸弃消化加入培养:弃去胰蛋白酶,注意更换吸管,加入新鲜的培养。【吹打分散细胞操作】1)吹打制悬:用滴管将已经消化细胞吹打成细胞悬;2)吸细胞悬入离心管:将细胞悬吸入10ml离心管中;3)平衡离心:平衡后将离心管放入台式离心机中,以1000转/分钟离心6-8分钟;4)弃上清,加入新培养:弃去上清,加入2ml培养,用滴管轻轻吹打细胞制成细胞悬。【分装稀释细胞操作】1)显微镜下观察细胞:倒置显微镜下观察细胞量,必要是计数。注意密度过小会影响传代细胞的生长,传代细胞的密度应该不低于5×105/ml。Zui后要做HAO标记;2)分装:将细胞悬吸出分装至2~3个培养瓶中,加入适量培养基旋紧瓶盖;3)继续培养:用酒精棉球擦拭培养瓶,适当旋松瓶盖,放入CO2培养箱中继续培养。传代细胞2小时后开始贴附在瓶壁上。
KYSE 140 Cells;背景说明:食管鳞癌细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:KU19-19细胞、SG231细胞、LC1/Sq细胞
NCI-SNU-1 Cells;背景说明:详见相关文献介绍;传代方法:2-3天换液1次。;生长特性:悬浮聚集;形态特性:上皮细胞;相关产品有:HCC-78细胞、MGSMC细胞、MH7A细胞
NCI-H1373 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HLMVEC细胞、Colon 38细胞、RIN Cl-5F细胞
231-luc Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:BTT739细胞、HCC-2185细胞、RPE1-hTERT细胞
LIM1215 Cells;背景说明:结直肠癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HCC44细胞、HL60细胞、MDAMB468细胞
Leukemic 1210 Cells;背景说明:该细胞源于用0.2%甲基胆蒽(溶解)涂抹雌性小鼠的皮肤诱发的肿瘤,鼠痘病毒阴性。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:NCI-H2108细胞、H-1693细胞、Co320细胞
MC-116 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2-3次。;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:SBC3细胞、NPA87-1细胞、H-510细胞
SF-763 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:CCRF SB细胞、RH8994细胞、L cell line细胞
GM00637B Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Neukoplast细胞、L-M(TK-)细胞、alphaTC1 Clone 6细胞
Spodoptera frugiperda clone 9 Cells;背景说明:详见相关文献介绍;传代方法:1:5传代;2-3天换液1次。;生长特性:混合生长;形态特性:上皮样;相关产品有:IPLB-Sf21细胞、VP 267细胞、KBM5细胞
DAN-G Cells;背景说明:胰腺癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:aNK细胞、MC 3T3-E1细胞、Ramos-2G6-4C10细胞
MA-c Cells;背景说明:星形胶质 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H69/P细胞、UMUC-3细胞、Alexander cells细胞
A.704 Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:4传代,每周换液2-3次;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:HEK293-EBNA细胞、16HBE140细胞、HSC-2细胞
TK 10 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:MC-116细胞、AG06814-N细胞、CAL851细胞
NCIH1869 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;每周换液2次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:H2.35细胞、NCI-H345细胞、NCI-H-295细胞
CCD-1112sk Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Renal Carcinoma细胞、Vertebral Cancer of the Prostate细胞、H727细胞
MDA-MB-134 VI Cells;背景说明:该细胞1973年由R. Cailleau建系,源自74岁乳腺导管癌女性患者的胸腔积液,细胞生长缓慢,松散贴壁,生长过程中会脱落到培养基,不会汇合,过表达FGF受体;传代方法:1:2—1:4传代,每周换液2—3次;生长特性:松散贴壁生长;形态特性:上皮细胞样;相关产品有:G292细胞、Centre Antoine Lacassagne-33细胞、Plaepi 34细胞
HT-29人结肠癌复苏细胞保种中心|带STR证书
Abcam HEK293T DAPK3 KO Cells(提供STR鉴定图谱)
AG09861 Cells(提供STR鉴定图谱)
BayGenomics ES cell line NPX269 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XC499 Cells(提供STR鉴定图谱)
BT-20/CMV-Luc Cells(提供STR鉴定图谱)
COS-1-InVitrus Cells(提供STR鉴定图谱)
DA04032 Cells(提供STR鉴定图谱)
DLD-1 ATG7(+/-) Cells(提供STR鉴定图谱)
GM03030 Cells(提供STR鉴定图谱)
HLFa Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代;每周换液2次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:H1563细胞、SNU-398细胞、H2196细胞
Mouse Forestomach Carcinoma Cells;背景说明:胃癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MCA 205细胞、GM00346B细胞、Capan-1细胞
COLO_320DM Cells;背景说明:该细胞可产生5-羟色胺、去甲、、ACTH和甲状旁腺素。角蛋白、波形蛋白弱阳性。培养条件: RPMI 1640 10%FBS;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮+贴壁;形态特性:淋巴细胞;相关产品有:HANK-1细胞、SUDHL-8细胞、H22细胞
H1581 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:混合型;形态特性:上皮样;相关产品有:HSC-6细胞、L 132细胞、SPDC-CCL141细胞
OVCA432 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KU 19-19细胞、REC-1细胞、H-838细胞
3-LL Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:CCD 966 SK细胞、SRS-82细胞、SW-954细胞
558-D100,10,1 Cells(提供STR鉴定图谱)
CAL 62 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NTC200细胞、TB1 Lu细胞、SP2-0-Ag14细胞
Lu-165 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HEY A8细胞、HuO9细胞、HCC 70细胞
BXPC3 Cells;背景说明:这个细胞株不表达囊肿性纤维化跨膜电导调节子(CFTR)。CFTR阳性的细胞株是Capan-1(ATCCHTB-79)。;传代方法:消化5-10分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:ARPE-19细胞、Roswell Park Memorial Institute 6666细胞、H-7721细胞
OCI Ly1 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCIH1437细胞、P3X63 Ag8.653细胞、SUDHL-2细胞
769-P Cells;背景说明:该细胞系1975年建系,源自一位63岁白人女性的初期透明细胞腺癌组织,细胞呈圆形且边界不清,核浆比大,有微绒毛及桥粒。该细胞可在软琼脂上生长。 ;传代方法:1:4—1:12传代,2—3天换液一次;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:143B细胞、MA 104细胞、RBE细胞
PANC0504 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:ATN1细胞、HIT细胞、SK-MEL-24细胞
3T3-F442A Cells;背景说明:脂肪前体细胞;雄性;Swiss albino;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:rRTEC细胞、SK-LU-1细胞、Panc 05.04细胞
DHL-8 Cells;背景说明:弥漫大B淋巴瘤;腹腔积液转移;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NS-1细胞、CAL-51细胞、PTK 2细胞
GM17687 Cells(提供STR鉴定图谱)
HAP1 HSPA1L (-) 2 Cells(提供STR鉴定图谱)
TSUpr1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:SNU761细胞、UCLA NPA871细胞、KG1细胞
NCI-HUT-596 Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:8传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:675T细胞、A 253细胞、CWR22Rv1细胞
2BS Cells;背景说明:胚肺;成纤维细胞;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:alphaTC1 Clone 6细胞、NCI-H2030细胞、H69细胞
SK-OV-3 Cells;背景说明:SK-OV-3由G.Trempe和L.J.Old在1973年从卵巢肿瘤病人的腹水分离得到。 此细胞对肿瘤坏死因子和几种细胞毒性药物包括白喉毒素、顺铂和阿霉素均耐受。 在裸鼠中致瘤,且形成与卵巢原位癌一致的中度分化的腺癌。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:上皮细胞样;相关产品有:WM-239-A细胞、TCMK1细胞、NCIH2405细胞
CAL 39 Cells;背景说明:外阴鳞癌细胞;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MM1S细胞、NCI-H1522细胞、MMAc.SF细胞
SUM-102 Cells;背景说明:乳腺癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-1651细胞、NRK 49F细胞、SKN-AS细胞
no-11 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:MiaPaca.2细胞、KMB-17细胞、HRIF细胞
KU19-19 Cells;背景说明:膀胱癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:LTEPa2细胞、GalK1细胞、PLB-985细胞
HPS0159 Cells(提供STR鉴定图谱)
JHU170i Cells(提供STR鉴定图谱)
MDA-BoM-1833 Cells(提供STR鉴定图谱)
ND33950 Cells(提供STR鉴定图谱)
PR01238 Cells(提供STR鉴定图谱)
U2OS ADRA1B DAG-Nomad Cells(提供STR鉴定图谱)
UMANe002-A-5 Cells(提供STR鉴定图谱)
HAP1 TSC2 (-) 1 Cells(提供STR鉴定图谱)
JM-1 Cells;背景说明:详见相关文献介绍;传代方法:换液2-3次一周;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:RPMI No. 1846细胞、Clone Y-1细胞、H7721细胞
COC1/CDDP Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:UCLA RO-81A-1细胞、IFRS1细胞、DHL-6细胞
MKN 45 Cells;背景说明:该细胞系由S Akiyama建立,源于一位35岁患有印戒细胞癌的女性的胃淋巴结。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁+悬浮;形态特性:淋巴母细胞;相关产品有:hRPTC细胞、SK-MEL-2-LUC细胞、T2细胞
Raji Cells;背景说明:Raji细胞由PulvertaftRJV于1963年从一位11岁黑人男孩的左上颌骨的Burkitt淋巴瘤中分离建立的,是第一个人类造血系统的连续传代细胞,为B细胞起源。该细胞中含有EBV,需要在二级生物安全柜中操作;可作转染宿主。;传代方法:维持细胞浓度在4×105/ml-3×106/ml;根据细胞浓度每2-3天补液1次。;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:VAESBJ细胞、Hs737T细胞、HGC-27细胞
251MG Cells;背景说明:U-251 MG分离至一位患者的胶质母细胞瘤组织。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:成纤维细胞样;相关产品有:Hs-578T细胞、IMCD-3细胞、HN 4细胞
251MG Cells;背景说明:U-251 MG分离至一位患者的胶质母细胞瘤组织。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:成纤维细胞样;相关产品有:Hs-578T细胞、IMCD-3细胞、HN 4细胞
H647ell Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;每周换液2次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:SMMC-7721细胞、C3H10T1/2细胞、OCIAML4细胞
OCIAML3 Cells;背景说明:急性髓系白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:BetaTC6细胞、GM2132细胞、HS-5细胞
V79-1 Cells;背景说明:肺;自发永生;雄性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MM96L细胞、Capan1细胞、HEY-A8细胞
CCD966SK Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KYSE-150细胞、Hs821T细胞、YD38细胞
231-luc Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:BTT739细胞、HCC-2185细胞、RPE1-hTERT细胞
231-luc Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:BTT739细胞、HCC-2185细胞、RPE1-hTERT细胞
MDA.MB.231 Cells;背景说明:MDA-MB-231来自患有转移乳腺腺癌的51岁女病人的胸水。在裸鼠和ALS处理的BALB/c小鼠中,它能形成低分化腺癌(III级)。;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:PANC 203细胞、VP267细胞、KY70细胞
ECV 304 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SK-MEL24细胞、NCI-H596细胞、THLE-2细胞
UM-UC-14 Cells;背景说明:肾癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CF-1 MEF细胞、GL261细胞、ZYM-DIEC02细胞
SKO-007 clone J3 Cells(提供STR鉴定图谱)
H1385 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MM.1S细胞、MD Anderson-Metastatic Breast-175-VIII细胞、HeLa229细胞
K299 Cells;背景说明:间变性大细胞淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:CL34细胞、H-2347细胞、NK-92细胞
Tregs Cells;背景说明:调节性T Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:Epstein-Barr-3细胞、SCC9细胞、SKNBE2细胞
293-H Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI/ADRRES细胞、SKNAS细胞、Vero E-6细胞
BNL CL2 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:BT 20细胞、MDA2B细胞、Hs-600-T细胞
Pa14C Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:DLD 1细胞、HL7702细胞、RPMI no. 8226细胞
U-87MG Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:D341Med细胞、B16/BL6细胞、TT细胞
SCL-II Cells;背景说明:皮肤鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:C41细胞、LL/2(LLc1)细胞、CCFSTTG1细胞
SNU182 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:U266细胞、CC-LP-1细胞、hTERT-RPE1细胞
Panc 05.04 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:KMST6细胞、C2C12细胞、U-251 MG细胞
NCIH1092 Cells;背景说明:详见相关文献介绍;传代方法:随细胞的密度而增加;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:RCC-JF细胞、SK_N_FI细胞、MHCCLM3细胞
UMNSAH-DF-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成纤维母细胞样;相关产品有:U343MG细胞、SW1116细胞、OSKM-1细胞
SVG p12 Cells;背景说明:星形胶质细胞;SV40转化;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H1993细胞、Clone Y1细胞、KU-812细胞
NCI-H211 Cells;背景说明:详见相关文献介绍;传代方法:3-4天换液1次。;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:H-1693细胞、RCC-4细胞、UMUC-3细胞
HT-29人结肠癌复苏细胞保种中心|带STR证书
BayGenomics ES cell line RRK254 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XL961 Cells(提供STR鉴定图谱)
Glut1(D4417-14) Cells(提供STR鉴定图谱)
OKT 8 Cells(提供STR鉴定图谱)
ZUE12 Cells(提供STR鉴定图谱)
MCW070i-40002330 Cells(提供STR鉴定图谱)
" "PubMed=327080; DOI=10.1093/jnci/59.1.221
Fogh J., Fogh J.M., Orfeo T.
One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice.
J. Natl. Cancer Inst. 59:221-226(1977)
PubMed=833871; DOI=10.1093/jnci/58.2.209
Fogh J., Wright W.C., Loveless J.D.
Absence of HeLa cell contamination in 169 cell lines derived from human tumors.
J. Natl. Cancer Inst. 58:209-214(1977)
PubMed=864752; DOI=10.1093/jnci/58.6.1743
Marshall C.J., Franks L.M., Carbonell A.W.
Markers of neoplastic transformation in epithelial cell lines derived from human carcinomas.
J. Natl. Cancer Inst. 58:1743-1751(1977)
PubMed=77569; DOI=10.1111/j.1399-0039.1978.tb01259.x
Espmark J.A., Ahlqvist-Roth L., Sarne L., Persson A.
Tissue typing of cells in culture. III. HLA antigens of established human cell lines. Attempts at typing by the mixed hemadsorption technique.
Tissue Antigens 11:279-286(1978)
PubMed=626984
Erickson L.C., Osieka R., Kohn K.W.
Differential repair of 1-(2-chloroethyl)-3-(4-methylcyclohexyl)-1- nitrosourea-induced DNA damage in two human colon tumor cell lines.
Cancer Res. 38:802-808(1978)
PubMed=468301; PMCID=PMC1457289
Trejdosiewicz L.K., Trejdosiewicz A.J., Ling N.R., Dykes P.W.
Growth enhancing property of human monocytes from normal donors and cancer patients.
Immunology 37:247-252(1979)
PubMed=6256643; DOI=10.1038/288724a0
Day R.S. 3rd, Ziolkowski C.H.J., Scudiero D.A., Meyer S.A., Lubiniecki A.S., Girardi A.J., Galloway S.M., Bynum G.D.
Defective repair of alkylated DNA by human tumour and SV40-transformed human cell strains.
Nature 288:724-727(1980)
PubMed=6445228
Kimball P.M., Brattain M.G.
Isolation of a cellular subpopulation from a human colonic carcinoma cell line.
Cancer Res. 40:1574-1579(1980)
PubMed=22282976; DOI=10.1093/carcin/1.1.21
Day R.S. 3rd, Ziolkowski C.H.J., Scudiero D.A., Meyer S.A., Mattern M.R.
Human tumor cell strains defective in the repair of alkylation damage.
Carcinogenesis 1:21-32(1980)
PubMed=7017212; DOI=10.1093/jnci/66.6.1003
Pollack M.S., Heagney S.D., Livingston P.O., Fogh J.
HLA-A, B, C and DR alloantigen expression on forty-six cultured human tumor cell lines.
J. Natl. Cancer Inst. 66:1003-1012(1981)
PubMed=7459858
Rousset M., Zweibaum A., Fogh J.
Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins.
Cancer Res. 41:1165-1170(1981)
PubMed=6220172
Dracopoli N.C., Fogh J.
Polymorphic enzyme analysis of cultured human tumor cell lines.
J. Natl. Cancer Inst. 70:469-476(1983)
PubMed=6825208; DOI=10.1093/carcin/4.2.199
Yarosh D.B., Foote R.S., Mitra S., Day R.S. 3rd
Repair of O6-methylguanine in DNA by demethylation is lacking in Mer- human tumor cell strains.
Carcinogenesis 4:199-205(1983)
PubMed=6500159; DOI=10.1159/000163283
Gershwin M.E., Lentz D., Owens R.B.
Relationship between karyotype of tissue culture lines and tumorigenicity in nude mice.
Exp. Cell Biol. 52:361-370(1984)
PubMed=6582512; DOI=10.1073/pnas.81.2.568; PMCID=PMC344720
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=3472642; DOI=10.1016/0165-4608(87)90267-6
Chen T.-R., Drabkowski D.J., Hay R.J., Macy M.L., Peterson W.D. Jr.
WiDr is a derivative of another colon adenocarcinoma cell line, HT-29.
Cancer Genet. Cytogenet. 27:125-134(1987)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=3349466
Chantret I., Barbat A., Dussaulx E., Brattain M.G., Zweibaum A.
Epithelial polarity, villin expression, and enterocytic differentiation of cultured human colon carcinoma cells: a survey of twenty cell lines.
Cancer Res. 48:1936-1942(1988)
PubMed=2288877
Hafez M.M., Infante D., Winawer S.J., Friedman E.
Transforming growth factor beta 1 acts as an autocrine-negative growth regulator in colon enterocytic differentiation but not in goblet cell maturation.
Cell Growth Differ. 1:617-626(1990)
PubMed=1778766; DOI=10.1111/j.1349-7006.1991.tb01816.x; PMCID=PMC5918361
Takeshima E., Hamaguchi M., Watanabe T., Akiyama S., Kataoka M., Ohnishi Y., Xiao H.-Y., Nagai Y., Takagi H.
Aberrant elevation of tyrosine-specific phosphorylation in human gastric cancer cells.
Jpn. J. Cancer Res. 82:1428-1435(1991)
PubMed=1937958; DOI=10.1002/ijc.2910490516
Lesuffleur T., Kornowski A., Luccioni C., Muleris M., Barbat A., Beaumatin J., Dussaulx E., Dutrillaux B., Zweibaum A.
Adaptation to 5-fluorouracil of the heterogeneous human colon tumor cell line HT-29 results in the selection of cells committed to differentiation.
Int. J. Cancer 49:721-730(1991)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=1389533; DOI=10.1016/0959-8049(92)90031-V
Lahm H., Petral-Malec D., Yilmaz-Ceyhan A., Fischer J.R., Lorenzoni M., Givel J.-C., Odartchenko N.
Growth stimulation of a human colorectal carcinoma cell line by interleukin-1 and -6 and antagonistic effects of transforming growth factor beta 1.
Eur. J. Cancer 28A:1894-1899(1992)
PubMed=1730571; DOI=10.1007/BF02631079
Goodwin T.J., Jessup J.M., Wolf D.A.
Morphologic differentiation of colon carcinoma cell lines HT-29 and HT-29KM in rotating-wall vessels.
In Vitro Cell. Dev. Biol. Anim. 28:47-60(1992)
PubMed=8464898; DOI=10.1073/pnas.90.7.2842; PMCID=PMC46192
Browning M.J., Krausa P., Rowan A.J., Bicknell D.C., Bodmer J.G., Bodmer W.F.
Tissue typing the HLA-A locus from genomic DNA by sequence-specific PCR: comparison of HLA genotype and surface expression on colorectal tumor cell lines.
Proc. Natl. Acad. Sci. U.S.A. 90:2842-2845(1993)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045; PMCID=PMC45163
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=7651727
Kastrinakis W.V., Ramchurren N., Rieger K.M., Hess D.T., Loda M., Steele G., Summerhayes I.C.
Increased incidence of p53 mutations is associated with hepatic metastasis in colorectal neoplastic progression.
Oncogene 11:647-652(1995)
PubMed=8895552; DOI=10.1002/(SICI)1097-0215(19960927)68:1<126::aid-ijc22>3.0.CO;2-8
Suardet L., Li C., Little J.B.
Radio-induced modulation of transforming growth factor beta1 sensitivity in a p53 wild-type human colorectal-cancer cell line.
Int. J. Cancer 68:126-131(1996)
PubMed=9000147
Cottu P.-H., Muzeau F., Estreicher A., Flejou J.-F., Iggo R.D., Thomas G., Hamelin R.
Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines.
Oncogene 13:2727-2730(1996)
PubMed=9000572
Hoang J.-M., Cottu P.-H., Thuille B., Salmon R.J., Thomas G., Hamelin R.
BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines.
Cancer Res. 57:300-303(1997)
PubMed=9294210; DOI=10.1073/pnas.94.19.10330; PMCID=PMC23362
Ilyas M., Tomlinson I.P.M., Rowan A.J., Pignatelli M., Bodmer W.F.
Beta-catenin mutations in cell lines established from human colorectal cancers.
Proc. Natl. Acad. Sci. U.S.A. 94:10330-10334(1997)
PubMed=10612807; DOI=10.1002/(SICI)1098-2264(200002)27:2<183::aid-gcc10>3.0.CO;2-P; PMCID=PMC4721570
Ghadimi B.M., Sackett D.L., Difilippantonio M.J., Schrock E., Neumann T., Jauho A., Auer G., Ried T.
Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations.
Genes Chromosomes Cancer 27:183-190(2000)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10737795; DOI=10.1073/pnas.97.7.3352; PMCID=PMC16243
Rowan A.J., Lamlum H., Ilyas M., Wheeler J.M.D., Straub J., Papadopoulou A., Bicknell D.C., Bodmer W.F., Tomlinson I.P.M.
APC mutations in sporadic colorectal tumors: a mutational 'hotspot' and interdependence of the 'two hits'.
Proc. Natl. Acad. Sci. U.S.A. 97:3352-3357(2000)
PubMed=11226274; DOI=10.1073/pnas.041603298; PMCID=PMC30173
Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D.C., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.W.
Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement.
Proc. Natl. Acad. Sci. U.S.A. 98:2538-2543(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11526487; DOI=10.1038/sj.onc.1204611
Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P.-H., Hamelin R.
Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines.
Oncogene 20:5025-5032(2001)
PubMed=11668190; DOI=10.1177/002215540104901105
Quentmeier H., Osborn M., Reinhardt J., Zaborski M., Drexler H.G.
Immunocytochemical analysis of cell lines derived from solid tumors.
J. Histochem. Cytochem. 49:1369-1378(2001)
PubMed=11687795; DOI=10.1038/ng754
Snijders A.M., Nowak N.J., Segraves R., Blackwood S., Brown N., Conroy J., Hamilton G., Hindle A.K., Huey B., Kimura K., Law S., Myambo K., Palmer J., Ylstra B., Yue J.P., Gray J.W., Jain A.N., Pinkel D., Albertson D.G.
Assembly of microarrays for genome-wide measurement of DNA copy number.
Nat. Genet. 29:263-264(2001)
PubMed=11921276; DOI=10.1002/gcc.10003
Kawai K., Viars C., Arden K.C., Tarin D., Urquidi V., Goodison S.
Comprehensive karyotyping of the HT-29 colon adenocarcinoma cell line.
Genes Chromosomes Cancer 34:1-8(2002)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=15748285; DOI=10.1186/1479-5876-3-11; PMCID=PMC555742
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. 3rd, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=15900046; DOI=10.1093/jnci/dji133
Mashima T., Oh-hara T., Sato S., Mochizuki M., Sugimoto Y., Yamazaki K., Hamada J.-i., Tada M., Moriuchi T., Ishikawa Y., Kato Y., Tomoda H., Yamori T., Tsuruo T.
p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target.
J. Natl. Cancer Inst. 97:765-777(2005)
PubMed=16083285; DOI=10.1021/pr050048h
Kim J.-E., Tannenbaum S.R., White F.M.
Global phosphoproteome of HT-29 human colon adenocarcinoma cells.
J. Proteome Res. 4:1339-1346(2005)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=18258742; DOI=10.1073/pnas.0712176105; PMCID=PMC2268141
Emaduddin M., Bicknell D.C., Bodmer W.F., Feller S.M.
Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells.
Proc. Natl. Acad. Sci. U.S.A. 105:2358-2362(2008)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921; PMCID=PMC4020356
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=19927377; DOI=10.1002/gcc.20730; PMCID=PMC2818350
Knutsen T., Padilla-Nash H.M., Wangsa D., Barenboim-Stapleton L., Camps J., McNeil N.E., Difilippantonio M.J., Ried T.
Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines.
Genes Chromosomes Cancer 49:204-223(2010)
PubMed=19941903; DOI=10.1016/j.jviromet.2009.11.022
Karger A., Bettin B., Lenk M., Mettenleiter T.C.
Rapid characterisation of cell cultures by matrix-assisted laser desorption/ionisation mass spectrometric typing.
J. Virol. Methods 164:116-121(2010)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192; PMCID=PMC2943514
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780; PMCID=PMC2920028
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=20831567; DOI=10.1111/j.1582-4934.2010.01170.x; PMCID=PMC3918049
Ma Y.-L., Zhang P., Wang F., Moyer M.P., Yang J.-J., Liu Z.-H., Peng J.-Y., Chen H.-Q., Zhou Y.-K., Liu W.-J., Qin H.-L.
Human embryonic stem cells and metastatic colorectal cancer cells shared the common endogenous human microRNA-26b.
J. Cell. Mol. Med. 15:1941-1954(2011)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21912889; DOI=10.1007/s10637-011-9744-z
Sutherland H.S., Hwang I.Y., Marshall E.S., Lindsay B.S., Denny W.A., Gilchrist C., Joseph W.R., Greenhalgh D., Richardson E., Kestell P., Ding A., Baguley B.C.
Therapeutic reactivation of mutant p53 protein by quinazoline derivatives.
Invest. New Drugs 30:2035-2045(2012)
PubMed=22336246; DOI=10.1016/j.bmc.2012.01.017
Kong D.-X., Yamori T.
JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs.
Bioorg. Med. Chem. 20:1947-1951(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096; PMCID=PMC3285665
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595; PMCID=PMC3526189
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66; PMCID=PMC3543849
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=22940288; DOI=10.1016/j.jsbmb.2012.08.003; PMCID=PMC3695570
Hobaus J., Fetahu I.S., Khorchide M., Manhardt T., Kallay E.
Epigenetic regulation of the 1,25-dihydroxyvitamin D3 24-hydroxylase (CYP24A1) in colon cancer cells.
J. Steroid Biochem. Mol. Biol. 136:296-299(2013)
PubMed=23546019; DOI=10.3892/ijo.2013.1868
Petitprez A., Poindessous V., Ouaret D., Regairaz M., Bastian G., Guerin E., Escargueil A.E., Larsen A.K.
Acquired irinotecan resistance is accompanied by stable modifications of cell cycle dynamics independent of MSI status.
Int. J. Oncol. 42:1644-1653(2013)
PubMed=23631600; DOI=10.1021/pr400260h
Loftus N.J., Lai L., Wilkinson R.W., Odedra R., Wilson I.D., Barnes A.J.
Global metabolite profiling of human colorectal cancer xenografts in mice using HPLC-MS/MS.
J. Proteome Res. 12:2980-2986(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342; PMCID=PMC4893961
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23932154; DOI=10.1016/j.radonc.2013.06.032
Salendo J., Spitzner M., Kramer F., Zhang X., Jo P., Wolff H.A., Kitz J., Kaulfuss S., Beissbarth T., belstein M., Ghadimi M., Grade M., Gaedcke J.
Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g.
Radiother. Oncol. 108:451-457(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35; PMCID=PMC3816225
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20; PMCID=PMC4178206
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25841592; DOI=10.1016/j.jprot.2015.03.019
Piersma S.R., Knol J.C., de Reus I., Labots M., Sampadi B.K., Pham T.V., Ishihama Y., Verheul H.M.W., Jimenez C.R.
Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines.
J. Proteomics 127:247-258(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=29787047; DOI=10.1007/978-3-319-16104-4_11
Martinez-Maqueda D., Miralles B., Recio I.
HT29 cell line.
(In book chapter) The impact of food bioactives on health. In vitro and ex vivo models; Verhoeckx K., Cotter P., Lopez-Exposito I., Kleiveland C., Lea T., Mackie A., Requena T., Swiatecka D., Wichers H. (eds.); pp.113-124; Springer; Cham; Switzerland (2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235; PMCID=PMC4762531
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52; PMCID=PMC4932877
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4; PMCID=PMC5087121
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=28179481; DOI=10.1158/1535-7163.MCT-16-0578
Tanaka N., Mashima T., Mizutani A., Sato A., Aoyama A., Gong B., Yoshida H., Muramatsu Y., Nakata K., Matsuura M., Katayama R., Nagayama S., Fujita N., Sugimoto Y., Seimiya H.
APC mutations as a potential biomarker for sensitivity to tankyrase inhibitors in colorectal cancer.
Mol. Cancer Ther. 16:752-762(2017)
PubMed=28192450; DOI=10.1371/journal.pone.0171435; PMCID=PMC5305277
Fasterius E., Raso C., Kennedy S.A., Rauch N., Lundin P., Kolch W., Uhlen M., Al-Khalili Szigyarto C.
A novel RNA sequencing data analysis method for cell line authentication.
PLoS ONE 12:E0171435-E0171435(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y; PMCID=PMC5498998
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010; PMCID=PMC5583477
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J.H., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29101300; DOI=10.15252/msb.20177701; PMCID=PMC5731344
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D.J., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29207035; DOI=10.3892/ijo.2017.4206
Olejniczak A., Szarynska M., Kmiec Z.
In vitro characterization of spheres derived from colorectal cancer cell lines.
Int. J. Oncol. 52:599-612(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023; PMCID=PMC7339254
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)
PubMed=32172478; DOI=10.1007/s12253-020-00805-3
Xu Y.-T., Zhang L., Wang Q.-L., Zheng M.-J.
Comparison of different colorectal cancer with liver metastases models using six colorectal cancer cell lines.
Pathol. Oncol. R"