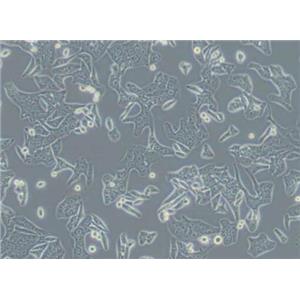
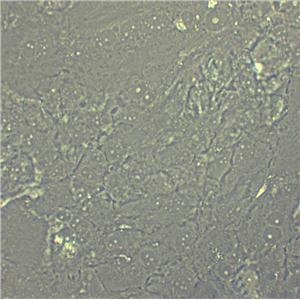
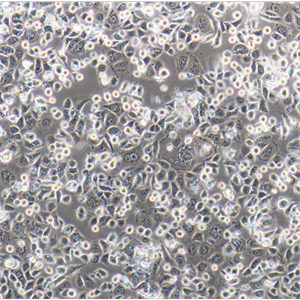
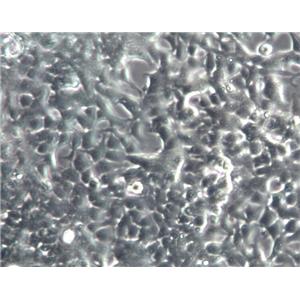
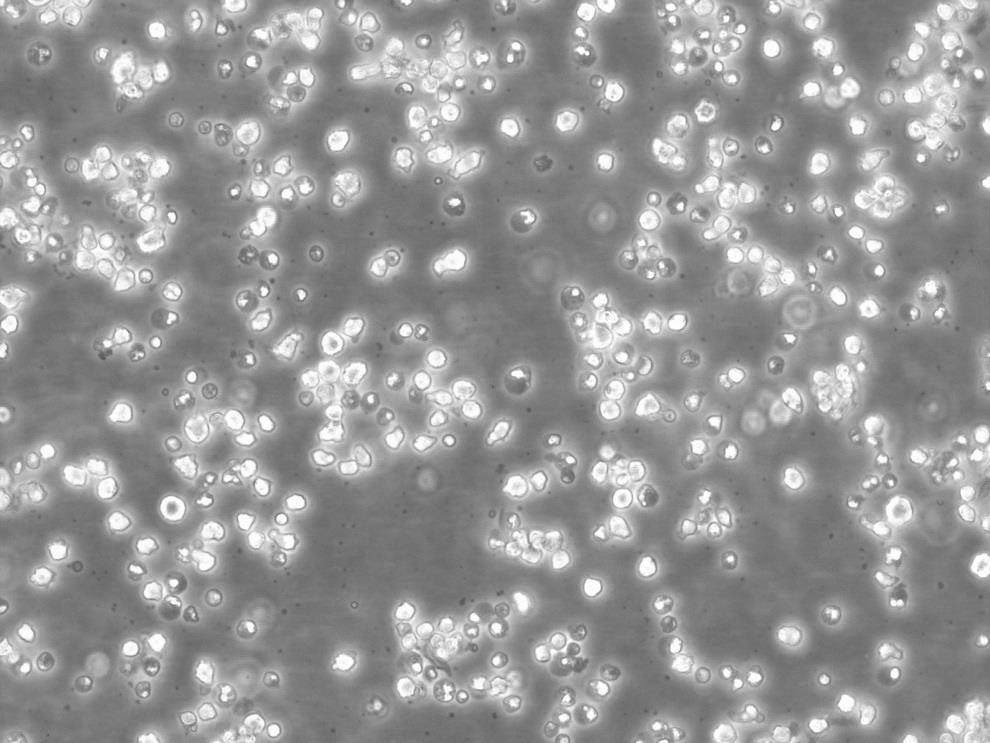
"SW620人结肠癌细胞全年复苏|已有STR图谱
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
公司细胞系形态漂亮、增殖倍数高、纯度高、功能性强,细胞培养就跟养孩子一个样。养孩子要喂奶,养细胞要加补液,都需要在前期补充足够的营养,初始状态的细胞或刚刚复苏的细胞还要适量加入血清或细胞因子来帮助它们的存活增殖,如果营养物质缺乏,细胞就会不生长甚至死亡。养孩子要从小培养学习,养细胞也得培养宝宝顺利生下来,你会经常抚摸他,给他看各种颜色,刺激他的五感。细胞也是一样,分离后的细胞需要使用特定的细胞因子进行活化、增殖。另外加入因子的种类、因子的浓度、加入时间、加入顺序都会影响细胞最终的结果。养孩子最怕孩子生病,养细胞最怕被污染,平时你会仔细观察宝宝是否呕吐、是否突然哭闹,猜测宝宝是否生病了。对于细胞,我们也需要时刻进行观察的,假如培养液浑浊(污染了),则需要换液后加抗生素;假如细胞增殖不明显,形态变差,则可能是因为营养不足了,对贴壁细胞可以消化后重新用新的培养基接种并加倍加入细胞因子含量;对悬浮细胞增殖能力不强的,则不着急补液,只是先补加血清、细胞因子看是否可以好转。培养时还得全程在无菌的环境,一个小小的偏差,细胞就会死亡。
换液周期:每周2-3次
KYSE-180 Cells;背景说明:食管鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SUM190细胞、PC-2 [Human pancreatic carcinoma]细胞、SKNBE(1)细胞
OLN 93 Cells;背景说明:胶质细胞;自发永生;Wistar;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Co320细胞、NCI-H2452细胞、Stanford University-Diffuse Histiocytic Lymphoma-16细胞
MIN6 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:NCIH1568细胞、10RGB细胞、LS 123细胞
背景信息:SW620是从一个51岁男性白人组织中分离得到。由A.Leibovitz等从一个淋巴结建株。细胞系主要由无绒毛的小园球细胞和双细胞组成。它仅合成少量癌胚抗原(A)且在裸鼠中有高度的致瘤性
SW620人结肠癌细胞全年复苏|已有STR图谱
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
DSMZ菌株保藏中心成立于1969年,是德国的国家菌种保藏中心。该中心一直致力于细菌、真菌、质粒、抗菌素、人体和动物细胞、植物病毒等的分类、鉴定和保藏工作。DSMZ菌种保藏中心是欧洲规模最大的生物资源中心,保藏有动物细胞500多株。Riken BRC成立于1920年,是英国的国家菌种保藏中心。该中心一直致力于细菌、真菌、植物病毒等的分类、鉴定和保藏工作。日本Riken BRC(Riken生物资源保藏中心)是全球三大典型培养物收集中心之一。Riken保藏中心提供了很多细胞系。在世界范围内,这些细胞系,都在医学、科学和兽医中具有重要意义。Riken生物资源中心支持了各种学术、健康、食品和兽医机构的研究工作,并在世界各地不同组织的微生物实验室和研究机构中使用。
COLO-1 Cells;背景说明:角质形成层;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs 766T细胞、OACP4 C细胞、Hs-675-T细胞
SK-MEL-28 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:8传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:星形的;相关产品有:HSF细胞、AQ-Mel细胞、4-1st细胞
H660 Cells;背景说明:详见相关文献介绍;传代方法:2-3天换液1次。;生长特性:悬浮生长;形态特性:上皮细胞;相关产品有:TE5细胞、MC3T3-E1(C4)细胞、UPCI-SCC-90细胞
1BH3 Cells(提供STR鉴定图谱)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
物种来源:人源、鼠源等其它物种来源
SW620人结肠癌细胞全年复苏|已有STR图谱
形态特性:上皮细胞样
细胞复苏后贴壁细胞较少的问题分析:总结1:复苏过程没有问题,是否是从拿出直接放入温水,还有培养箱,二氧化碳浓度,培养基、PH值等环节。要么加GAO浓度FBS 15-20%,看看能否帮助贴壁,当然也需要考虑血清问题,还有确信拿来的细胞没问题。总结2:首先应该怀疑冻存,实际上复苏出问题的可能非常小,因为操作简单,而且死板。1、你冻存的时候是不是消化的时间过长,这是一般人所注意不到的,即使书上也不讲这个问题,太长的消化时间会让细胞复苏时失去贴壁能力,表现为先贴后死,原因是在你复苏的时候细胞已进入凋亡程序,不可逆转的死亡。2、你的冻存HAO不HAO,是什么,甘油还是DMSO,质量非常重要,否则也会死亡。3、你的冻存的量加的是不是太多,AC推荐是不超过7%,大于5%,太多也不HAO。4、你在冻存的时候是不是把DMSO混均匀,这个有一些影响,但不算太大。5、你的冻存是否按部就班,就是所温度梯度是不是把握严格,很多人容易忘却这个事情,因为这个东西流程长。6、如果你细胞污染,你是否能很快看到,我比我的导师能早一天看到污染。从这个角度讲建议去除离心这步。7、你的细胞在冻存前是否过密。还有,不赞成孵箱污染这个概念的,所有在一个孵箱里的细胞都污染一个细菌的话,这个细菌是源于孵箱的,但这不代表孵箱污染,因为孵箱无论你如何处理都有大量的细菌,问题在操作。每次污染的原因都要尽可能的找,以后就不犯同样的问题,这个很重要,不能靠猜,否则你就有可能细胞养绝Zui后换课题,这个见得太多了,别不当会事。
MHHCALL2 Cells;背景说明:急性B淋巴细胞白血病;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:SK-MEL-24细胞、Mouse Forestomach Carcinoma细胞、WM35细胞
HEC1-B Cells;背景说明:该细胞是H.Kuramoto1968年分离的HEC-1-A细胞亚株。不同於HEC-A-1的是:该亚株在培养第135天到190天之间表现出稳定的生长周期,且重现扁平,与亲本细胞系相比更具铺路石式样。此外主要染色体组是亲本细胞的两倍。;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HConEpic细胞、HT 1376.T细胞、GDM-1细胞
RERF-LC-MS Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:NH6细胞、H-211细胞、RBL-I细胞
JIII Cells;背景说明:单核细胞白血病;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:MDA-MB-468细胞、LS1034细胞、3D4/21细胞
NCI-H498 Cells;背景说明:结肠腺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:半贴壁;形态特性:详见产品说明书;相关产品有:RPE1-hTERT细胞、B16 subline B78细胞、22Rv-1细胞
A549ATCC Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H-1435细胞、SJCRH30细胞、RGB细胞
Y79 Cells;背景说明:1971年1月,该细胞由ReidTW及其同事从病人右眼切除的肿瘤进行原代培养建立而成,此病人有很强的视网膜母细胞瘤的母系家族遗传性。该细胞的超微结构,如核膜内折、三层膜结构、大的被膜小泡、环孔板、微管、中心粒、基粒等都与原始肿瘤相似。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:圆形,成簇生长;相关产品有:CHL-1细胞、MAVER细胞、NCI-H211细胞
H-2122 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:淋巴母细胞;相关产品有:SF126细胞、MDA231-LM2-4175细胞、TF1细胞
J 111 Cells;背景说明:单核细胞白血病;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCI-H2030细胞、MRMT-1细胞、L929细胞
RINm-5F Cells;背景说明:胰岛β细胞瘤;雄性;NEDH;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Capan-2细胞、NCIH1781细胞、hs 68细胞
MPP 89 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KY180细胞、Sp 2/0-Ag 14细胞、INS1细胞
NCI-H23 Cells;背景说明:详见相关文献介绍;传代方法:1:3传代;3-4天1次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HEL-92细胞、H676细胞、VSMC细胞
CFPAC Cells;背景说明:详见相关文献介绍;传代方法:1:3-10传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:NCIH524细胞、HEL9217细胞、DMS-153细胞
CMT167 Cells;背景说明:肺腺癌;雌性;C57BL/ICRF-a(t);传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NS1/1-Ag4.1细胞、LIM-1215细胞、MC/CAR细胞
Tb1.Lu Cells;背景说明:肺;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CPA47细胞、H-1869细胞、L363细胞
SNU878 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:L cell细胞、CMECs细胞、A-875细胞
Karpas 422 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:WC00059细胞、NS-1细胞、TC7细胞
A-1632 Cells(提供STR鉴定图谱)
Abcam MCF-7 NCR1 KO Cells(提供STR鉴定图谱)
AVP-Ser18del-iPSC Cells(提供STR鉴定图谱)
BayGenomics ES cell line RRO326 Cells(提供STR鉴定图谱)
BayGenomics ES cell line YHB390 Cells(提供STR鉴定图谱)
CCSP C2 Cells(提供STR鉴定图谱)
DA01160 Cells(提供STR鉴定图谱)
DA05344 Cells(提供STR鉴定图谱)
GENEA071 Cells(提供STR鉴定图谱)
RCCJF Cells;背景说明:肾透明细胞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HSC-1细胞、UMUC1细胞、CLONE M3细胞
ABC-1 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:NCIH1869细胞、L 363细胞、NCIH1930细胞
OCIAML5 Cells;背景说明:急性髓系白血病细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:Leukemia 1210细胞、HS-294细胞、HEL-92-1-7细胞
Molm 14 Cells;背景说明:急性髓系白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:M-O7e细胞、HCO细胞、38C-13细胞
SCL-II Cells;背景说明:皮肤鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:C41细胞、LL/2(LLc1)细胞、CCFSTTG1细胞
Calu3 Cells;背景说明:该细胞是从一名25岁的白人男性肺腺癌患者的胸水中分离建立的;该患者曾使用过环磷酰胺、博来霉素、阿霉素进行治疗。该细胞接种至裸鼠可成瘤;可作转染宿主。;传代方法:消化20分钟。1:2。5-6天长满。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:PY8119细胞、CV1细胞、TM3细胞
KYSE-30 Cells;背景说明:来源于一位64岁,患有高分化的中段食管鳞癌的男性患者。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:上皮细胞样;相关产品有:95-D细胞、NCTC929细胞、3T3 L1细胞
C-33A Cells;背景说明:C-33A细胞株是N. Auersperg从宫颈癌切片中建立的一系列细胞株(参见ATCC CRL-1594和ATCC CRL-1595)中的一株。 细胞一开始就表现出亚二倍体核型及上皮细胞形态。 连续传代可以观察到核型不稳定。 存在成视网膜细胞瘤蛋白(pRB),但大小不正常。 P53表达上调,且有一个273位密码子的点突变导致Arg -> Cys的替换。 人乳头瘤病毒DNA及RNA阴性。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:NCIH740细胞、CEM-T4细胞、PC3M-IE8细胞
SW620人结肠癌细胞全年复苏|已有STR图谱
NCI-SNU-C1 Cells;背景说明:详见相关文献介绍;传代方法:每3-5天换液。;生长特性:悬浮生长;形态特性:上皮细胞;相关产品有:SH-SY5Y细胞、HeLa S3细胞、PC-9/GR细胞
NCIH727 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:NCI-N87细胞、TN5B14细胞、SW1573细胞
NCIH2342 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:6传代 ;生长特性:贴壁生长;形态特性:上皮样;相关产品有:RTMC细胞、ssMCF-7细胞、CAL148细胞
C32 [Human melanoma] Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代,2天换液1次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:T/GHA-VSMC细胞、WC00097细胞、SCC-15细胞
B16 Cells;背景说明:该细胞源于C57BL/6J小鼠黑色素瘤,可以产生黑色素,同基因小鼠体内移植可成瘤;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:梭形;相关产品有:Panc-08.13细胞、CCD33Co细胞、Panc-8_13细胞
B16 Cells;背景说明:该细胞源于C57BL/6J小鼠黑色素瘤,可以产生黑色素,同基因小鼠体内移植可成瘤;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:梭形;相关产品有:Panc-08.13细胞、CCD33Co细胞、Panc-8_13细胞
L(TK-) Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MC57G细胞、SUNE-1细胞、RKO-AS-45-1细胞
GM09723 Cells(提供STR鉴定图谱)
HAP1 BAX (-) 2 Cells(提供STR鉴定图谱)
H-1581 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:混合型;形态特性:上皮样;相关产品有:NCI-H841细胞、HLE细胞、YH细胞
NS-20Y Cells;背景说明:神经母细胞瘤;雄性;A/J;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CCRF CEM细胞、VK2细胞、U2932细胞
Jijoye Cells;背景说明:详见相关文献介绍;传代方法:每周2-3次。;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:D283细胞、SRA 01/04细胞、Mo 59J细胞
PFSK-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:GT38细胞、Tu-212细胞、Roswell Park Memorial Institute 8402细胞
RC-2 Cells;背景说明:来源于日本人的肾脏肿瘤细胞。 可以移植到裸鼠。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:HR-1细胞、HPDE细胞、OKT3细胞
HBE Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:NCI-446细胞、RL细胞、mIMCD3细胞
MD Anderson-Metastatic Breast-436 Cells;背景说明:该细胞源于一名43岁患有乳腺腺癌女性的胸腔积液。;传代方法:1:2传代,每周换液2—3次;生长特性:贴壁生长;形态特性:多角形;相关产品有:GLRK 13细胞、EJ1细胞、HTR-8/SVneo细胞
MCF 10A Cells;背景说明:乳腺;上皮细胞;自发永生;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:OCI-Ly01细胞、HBdSMC细胞、University of Michigan-Renal Carcinoma-2细胞
HG03140 Cells(提供STR鉴定图谱)
IBMS-iPSC-084-01 Cells(提供STR鉴定图谱)
LM-MEL-82 Cells(提供STR鉴定图谱)
NCI-H1628 Cells(提供STR鉴定图谱)
PathHunter CHO-K1 mGHSR1a beta-arrestin Cells(提供STR鉴定图谱)
Ubigene A-549 CACNA1D KO Cells(提供STR鉴定图谱)
UoC-B7 Cells(提供STR鉴定图谱)
HG02351 Cells(提供STR鉴定图谱)
VSMC Cells;背景说明:胸主动脉平滑肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hce8693细胞、Calu3细胞、IGROV 1细胞
NRK clone 52E Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:PBL细胞、ARO81细胞、NIH:OVCAR-5细胞
H-1373 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HCO细胞、CHP 212细胞、MCFs细胞
C4-1 Cells;背景说明:宫颈鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:IOSE80UBC细胞、HeLa S 3细胞、COR-L88细胞
K1735 Cells;背景说明:黑色素瘤; C3H/HeN;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:C3H 10T1/2细胞、Hs 742.T细胞、LI7细胞
K1735 Cells;背景说明:黑色素瘤; C3H/HeN;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:C3H 10T1/2细胞、Hs 742.T细胞、LI7细胞
GA-10(Clone 4) Cells;背景说明:详见相关文献介绍;传代方法:每周换液两次;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:hTERTHME1细胞、UCH1细胞、MKN-45细胞
3T3-L1 ad Cells;背景说明:3T3-L1是从3T3细胞(Swissalbino)中经克隆分离得到的连续传代的亚系。该细胞从快速分裂到汇合和接触性抑制状态经历了前脂肪细胞到脂肪样细胞的转变。该细胞鼠痘病毒阴性;可产生甘油三酯,高浓度血清可增强细胞内脂肪堆积。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:D-283 Med细胞、Hs 819.T细胞、H2009细胞
LoVo Cells;背景说明:LoVo建自1971年,从诊断为结肠腺癌的56岁男性白人的一个转移到左锁骨上区的肿瘤结节建系。癌基因C-myc,K-ras,H-ras,N-ras,Myb,sis和fos的表达呈阳性。癌基因N-myc的表达未做检测。它在裸鼠中能成瘤。与107细胞联合皮下接种5只裸鼠21天后全部成瘤。表达肿瘤特异的核基质蛋白蛋白CC-3和CC-4。;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:KYSE 270细胞、SNU878细胞、NCI-H1105细胞
P30 OHK Cells;背景说明:详见相关文献介绍;传代方法:10^5 cells/60mm dish;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:Ly1细胞、M-1细胞、SK-MEL-31细胞
Jurkat clone A3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LL/2 (LLC1)细胞、RTMC细胞、RIN m5F细胞
Karpas 422 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:WC00059细胞、NS-1细胞、TC7细胞
FTC133 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:HCC1395细胞、HSAS1细胞、HCA 7细胞
MPASMC Cells;背景说明:肺动脉平滑肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Panc_03_27细胞、H-1672细胞、T-HSC细胞
MUTZ3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:Vero-76细胞、HCC202细胞、HUCCT1细胞
SA085 Cells(提供STR鉴定图谱)
FL-83B Cells;背景说明:肝;自发永生;C57BL/6J;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SW954细胞、Hi-5细胞、MDA175细胞
CHO K1 Cells;背景说明:1957年,PuckTT从成年中国仓鼠卵巢的活检组织建立了CHO细胞,CHO-K1是CHO的一个亚克隆。CHO-K1的生长需要脯酸。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:CAL 78细胞、NCIH1623细胞、NCI-H82细胞
Mc Ardle 7777 Cells;背景说明:肝癌;雌性;Buffalo;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:PLC-PRF-5细胞、MBMEC细胞、SUDHL-6细胞
HUVEC-C[HUVEC] Cells;背景说明:脐静脉;内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:N1S1细胞、M-07e细胞、Virginia Mason Research Center-Lung Cancer D细胞
H838 Cells;背景说明:该细胞于1984年建系,源于一位59岁患有非小细胞肺癌的白人男性吸烟者,从患者淋巴结转移灶分离而来。;传代方法:1:3-1:6传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:E.G7-OVA细胞、Hs 600.T细胞、AML-2细胞
HIT T15 Cells;背景说明:胰岛β细胞;SV40转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SuDHL 8细胞、DH82细胞、TCMK-1细胞
NCTC929 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:OAW-42细胞、GM07404细胞、SK-N-BE(1)n细胞
IHH Cells;背景说明:肝;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HS5细胞、Jurkat-E6细胞、LPC-H12细胞
SW620人结肠癌细胞全年复苏|已有STR图谱
MDCK-2 Cells;背景说明:详见相关文献介绍;传代方法:1:3传代,3-4天传1次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Ku812F细胞、HS578T细胞、Swiss/3T3细胞
KMH-2 Cells;背景说明:未分化甲状腺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:OCI-Ly1细胞、Epithelioma Papulosum Cyprini细胞、EAhy926细胞
SUDHL8 Cells;背景说明:弥漫大B淋巴瘤;腹腔积液转移;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:OCI Ly19细胞、MOLT 3细胞、MDA 1386细胞
U-118-MG Cells;背景说明:注意: 据报道来自不同个体的胶质母细胞瘤细胞株U-118 MG (HTB-15) 和 U-138 MG (HTB-16)有着一致的VNTR和相近的STR模式。 U-118 MG 和 U-138 MG细胞遗传学上很相似并有至少六个衍生标记染色体。 这是1966年至1969年间J. Ponten和同事从恶性神经胶质瘤中构建的细胞株中的一株(其它包括ATCC HTB-14和 ATCC HTB-16 and ATCC HTB-17)。 1987年用BM-Cycline培养6周去除了支原体污染。 ;传代方法: 消化3-5分钟。1:2传代。3天内可长满。;生长特性:贴壁生长;形态特性:混合型;相关产品有:KP4细胞、CaSki细胞、NCIH716细胞
Vx-2 Cells;背景说明:表皮鳞癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:GM17219细胞、SupB15W细胞、HEC1A细胞
H4IIEC3 Cells;背景说明:在糖皮质激素、胰岛素或cAMP衍生物的诱导下可以产生酪酸基转移酶;可被逆转录病毒感染;可产生白蛋白、转铁蛋白、凝血酶原;在AxC大鼠中可以成瘤。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:OCI-LY-7细胞、H1792细胞、Hs729细胞
BayGenomics ES cell line RRI191 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XH679 Cells(提供STR鉴定图谱)
F121-9 Cells(提供STR鉴定图谱)
NCI-HPN-F4K Colon Cells(提供STR鉴定图谱)
UV.BAL-1.1 Cells(提供STR鉴定图谱)
MCC148c Cells(提供STR鉴定图谱)
" "PubMed=924690; DOI=10.1002/ijc.2910200505
Kerbel R.S., Pross H.F., Leibovitz A.
Analysis of established human carcinoma cell lines for lymphoreticular-associated membrane receptors.
Int. J. Cancer 20:673-679(1977)
PubMed=286328; DOI=10.1073/pnas.76.3.1438; PMCID=PMC383267
Herlyn M., Steplewski Z., Herlyn D., Koprowski H.
Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 76:1438-1442(1979)
PubMed=288927; DOI=10.1093/jnci/63.3.635
Leibovitz A., Wright W.C., Pathak S., Siciliano M.J., Daniels W.P., Fogh H., Fogh J.
Detection and analysis of a glucose 6-phosphate dehydrogenase phenotype B cell line contamination.
J. Natl. Cancer Inst. 63:635-645(1979)
PubMed=6582512; DOI=10.1073/pnas.81.2.568; PMCID=PMC344720
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045; PMCID=PMC45163
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=9023415; DOI=10.1006/cimm.1996.1062
Seki N., Hoshino T., Kikuchi M., Hayashi A., Itoh K.
HLA-A locus-restricted and tumor-specific CTLs in tumor-infiltrating lymphocytes of patients with non-small cell lung cancer.
Cell. Immunol. 175:101-110(1997)
PubMed=9178645; DOI=10.1006/cimm.1997.1108
Nakao M., Sata M., Saitsu H., Yutani S., Kawamoto M., Kojiro M., Itoh K.
CD4+ hepatic cancer-specific cytotoxic T lymphocytes in patients with hepatocellular carcinoma.
Cell. Immunol. 177:176-181(1997)
PubMed=9290701; DOI=10.1002/(SICI)1098-2744(199708)19:4<243::aid-mc5>3.0.CO;2-D
Jia L.-Q., Osada M., Ishioka C., Gamo M., Ikawa S., Suzuki T., Shimodaira H., Niitani T., Kudo T., Akiyama M., Kimura N., Matsuo M., Mizusawa H., Tanaka N., Koyama H., Namba M., Kanamaru R., Kuroki T.
Screening the p53 status of human cell lines using a yeast functional assay.
Mol. Carcinog. 19:243-253(1997)
PubMed=9294210; DOI=10.1073/pnas.94.19.10330; PMCID=PMC23362
Ilyas M., Tomlinson I.P.M., Rowan A.J., Pignatelli M., Bodmer W.F.
Beta-catenin mutations in cell lines established from human colorectal cancers.
Proc. Natl. Acad. Sci. U.S.A. 94:10330-10334(1997)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10773689; DOI=10.1159/000015508
Melcher R., Steinlein C., Feichtinger W., Muller C.R., Menzel T., Luehrs H., Scheppach W., Schmid M.
Spectral karyotyping of the human colon cancer cell lines SW480 and SW620.
Cytogenet. Cell Genet. 88:145-152(2000)
PubMed=11113861; DOI=10.1002/1096-9896(2000)9999:9999<::aid-path775>3.0.CO;2-K
Hewitt R.E., McMarlin A., Kleiner D.E., Wersto R.P., Martin P., Tsokos M.G., Stamp G.W.H., Stetler-Stevenson W.G.
Validation of a model of colon cancer progression.
J. Pathol. 192:446-454(2000)
PubMed=11226274; DOI=10.1073/pnas.041603298; PMCID=PMC30173
Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D.C., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.W.
Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement.
Proc. Natl. Acad. Sci. U.S.A. 98:2538-2543(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12107847; DOI=10.1038/sj.bjc.6600459; PMCID=PMC2376111
Sekhon J., Pereira P.A., Sabbaghian N., Schievella A.R., Rozen R.
Antisense inhibition of methylenetetrahydrofolate reductase reduces survival of methionine-dependent tumour lines.
Br. J. Cancer 87:225-230(2002)
PubMed=15748285; DOI=10.1186/1479-5876-3-11; PMCID=PMC555742
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. 3rd, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=16854228; DOI=10.1186/1476-4598-5-29; PMCID=PMC1550420
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=18258742; DOI=10.1073/pnas.0712176105; PMCID=PMC2268141
Emaduddin M., Bicknell D.C., Bodmer W.F., Feller S.M.
Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells.
Proc. Natl. Acad. Sci. U.S.A. 105:2358-2362(2008)
PubMed=19188929; DOI=10.1038/tpj.2008.20
Guo J., Anderson M.G., Tapang P., Palma J.P., Rodriguez L.E., Niquette A.L., Li J.-L., Bouska J.J., Wang G., Semizarov D., Albert D.H., Donawho C.K., Glaser K.B., Shah O.J.
Identification of genes that confer tumor cell resistance to the aurora B kinase inhibitor, AZD1152.
Pharmacogenomics J. 9:90-102(2009)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921; PMCID=PMC4020356
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=19927377; DOI=10.1002/gcc.20730; PMCID=PMC2818350
Knutsen T., Padilla-Nash H.M., Wangsa D., Barenboim-Stapleton L., Camps J., McNeil N.E., Difilippantonio M.J., Ried T.
Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines.
Genes Chromosomes Cancer 49:204-223(2010)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192; PMCID=PMC2943514
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780; PMCID=PMC2920028
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21912889; DOI=10.1007/s10637-011-9744-z
Sutherland H.S., Hwang I.Y., Marshall E.S., Lindsay B.S., Denny W.A., Gilchrist C., Joseph W.R., Greenhalgh D., Richardson E., Kestell P., Ding A., Baguley B.C.
Therapeutic reactivation of mutant p53 protein by quinazoline derivatives.
Invest. New Drugs 30:2035-2045(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096; PMCID=PMC3285665
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595; PMCID=PMC3526189
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66; PMCID=PMC3543849
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=23631600; DOI=10.1021/pr400260h
Loftus N.J., Lai L., Wilkinson R.W., Odedra R., Wilson I.D., Barnes A.J.
Global metabolite profiling of human colorectal cancer xenografts in mice using HPLC-MS/MS.
J. Proteome Res. 12:2980-2986(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342; PMCID=PMC4893961
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23932154; DOI=10.1016/j.radonc.2013.06.032
Salendo J., Spitzner M., Kramer F., Zhang X., Jo P., Wolff H.A., Kitz J., Kaulfuss S., Beissbarth T., belstein M., Ghadimi M., Grade M., Gaedcke J.
Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g.
Radiother. Oncol. 108:451-457(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35; PMCID=PMC3816225
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20; PMCID=PMC4178206
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=24840470; DOI=10.1016/j.jprot.2014.05.002
Chik J.H.L., Zhou J., Moh E.S.X., Christopherson R., Clarke S.J., Molloy M.P., Packer N.H.
Comprehensive glycomics comparison between colon cancer cell cultures and tumours: implications for biomarker studies.
J. Proteomics 108:146-162(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235; PMCID=PMC4762531
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52; PMCID=PMC4932877
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27470641; DOI=10.1021/acs.jproteome.6b00391
Guo J.-H., Cui Y.-Z., Yan Z.-Q., Luo Y.-Z., Zhang W.-L., Deng S.-Y., Tang S.-Q., Zhang G., He Q.-Y., Wang T.
Phosphoproteome characterization of human colorectal cancer SW620 cell-derived exosomes and new phosphosite discovery for C-HPP.
J. Proteome Res. 15:4060-4072(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4; PMCID=PMC5087121
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=27987026; DOI=10.1007/s00216-016-0125-5; PMCID=PMC5303640
Schunter A.J., Yue X.-S., Hummon A.B.
Phosphoproteomics of colon cancer metastasis: comparative mass spectrometric analysis of the isogenic primary and metastatic cell lines SW480 and SW620.
Anal. Bioanal. Chem. 409:1749-1763(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y; PMCID=PMC5498998
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010; PMCID=PMC5583477
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J.H., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29101300; DOI=10.15252/msb.20177701; PMCID=PMC5731344
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D.J., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29131639; DOI=10.1021/acs.jproteome.7b00548
Torres S., Garcia-Palmero I., Marin-Vicente C., Bartolome R.A., Calvino E., Fernandez-Acenero M.J., Casal J.I.
Proteomic characterization of transcription and splicing factors associated with a metastatic phenotype in colorectal cancer.
J. Proteome Res. 17:252-264(2018)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051; PMCID=PMC6343826
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023; PMCID=PMC7339254
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)"