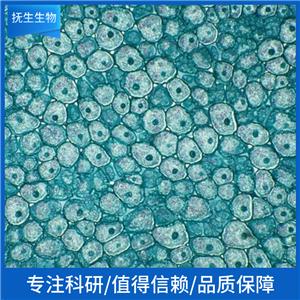
"SW480[SW-480]人结肠癌细胞全年复苏|已有STR图谱
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
【细胞培养经验分享】启蒙老师的重要性:一般进实验室都有师兄师姐带着做,他们就是你做细胞的启蒙老师。他们的操作手法、细节、理论讲解就成了你操作的准则,如营养液、细胞瓶的摆放位置、灭菌处理程序、开盖手法、细胞吹打手法等等。要学会他们的正确操作,在第一次的时候就要重视。像养孩子一样养细胞,细胞有时真的很脆弱,最好每天都去看看它,以防止出现培养箱缺水、缺二氧化碳、停电、温度不够等异常现象,也好及时解决这些意外,避免重复实验带来的更大痛苦。好细胞要及时保种:细胞要分批传代,这样即使有一批出了问题,还有一批备用的。像后者一般人可能不容易做到。但这是我血的教训,有一次细胞污染了,全军覆没。当时可后悔没有保种。细胞跟人一样,不同的细胞,培养特性是不一样的。培养过程中要细细体会,不同细胞系使用不同的培养基和血清。
换液周期:每周2-3次
NW38 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H1648细胞、MDA MB 361细胞、NTERA2-D1细胞
Mel-MeWo Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:5传代,2-3天换液1次。;生长特性:混合生长;形态特性:成纤维细胞;相关产品有:UK Pan-1细胞、K562细胞、P3HR1-BL细胞
Human Kidney-2 Cells;背景说明:该细胞属源于正常肾的近曲小管细胞,通过导入HPV-16 E6/E7基因而获得永生化。将含有HPV-16 E6/E7基因的重组的逆转录病毒载体pLXSN 16 E6/E7转染外生包装细胞Psi-2,Psi-2细胞产生的病毒再去感染兼嗜性包装细胞系PA317,最后将PA317产生的病毒颗粒导入正常的肾皮质近曲小管细胞。尽管pLXSN 16 E6/E7中含有新霉素抗性,但未用G418筛选转导克隆。Southern和FISH分析显示HK-2细胞来源于单克隆。PCR检测证实HK-2细胞基因组中含有E6/E7基因。;传代方法:1:4传代;2-3天换液1次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:NCI-H1975细胞、B16-F10细胞、H-1876细胞
背景信息:SW480源自一位51岁白人男性患者的原位直肠腺癌,而SW620源自同一病人一年后的淋巴结转移灶。该细胞C和直肠抗原3阴性;角蛋白阳性;p53基因第273位密码子的G→A突变引起Arg→His替代,309位密码子的C→T突变导致Pro→Ser替代;细胞p53蛋白表达水平升高;癌基因c-myc、K-ras、H-ras、N-ras、myb、sis和fos的表达呈阳性;未检测到癌基因N-myc的表达;不表达Matrilysin(一种与肿瘤侵袭相关的金属蛋白酶)。
SW480[SW-480]人结肠癌细胞全年复苏|已有STR图谱
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
DSMZ菌株保藏中心成立于1969年,是德国的国家菌种保藏中心。该中心一直致力于细菌、真菌、质粒、抗菌素、人体和动物细胞、植物病毒等的分类、鉴定和保藏工作。DSMZ菌种保藏中心是欧洲规模最大的生物资源中心,保藏有动物细胞500多株。Riken BRC成立于1920年,是英国的国家菌种保藏中心。该中心一直致力于细菌、真菌、植物病毒等的分类、鉴定和保藏工作。日本Riken BRC(Riken生物资源保藏中心)是全球三大典型培养物收集中心之一。Riken保藏中心提供了很多细胞系。在世界范围内,这些细胞系,都在医学、科学和兽医中具有重要意义。Riken生物资源中心支持了各种学术、健康、食品和兽医机构的研究工作,并在世界各地不同组织的微生物实验室和研究机构中使用。
BGC823 Cells;背景说明:建自一位62岁的胃癌患者;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:KM12SM细胞、Hep G2/C3A细胞、MLMA细胞
XWLC-05 Cells;背景说明:肺癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H1770细胞、V79-1细胞、Hela-mock细胞
MCF10-A Cells;背景说明:乳腺;上皮细胞;自发永生;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:ME-1 [Human leukemia]细胞、SNU387细胞、SNU251细胞
24-2-3 Cells(提供STR鉴定图谱)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
物种来源:人源、鼠源等其它物种来源
SW480[SW-480]人结肠癌细胞全年复苏|已有STR图谱
形态特性:上皮细胞样
细胞冻存复苏材料与方法步骤:常用的细胞冷冻贮存器为贮存器,规格有35L和50L两种。使用时要注意以下几点:(1)一般两周需充一次,至少一个月充一次。温度达-196℃,使用时注意勿让溅到皮肤上,以免引起冻伤。(2)容器为双层结构,中间为真空层,瓶口有双层焊接处,应防止焊接部裂开。(3)在装入时,要注意缓慢小心,并用厚纸卷筒或制漏斗作引导,使直达瓶底,如有专用灌注装置则更HAO。若为初次使用,加时更要缓慢,以免温度骤降而使容器损坏。细胞冻存时常备的材料有:0.25%胰蛋白酶,含10%~20%的血清培养,DMSO(分析纯)或无色新鲜甘油(121°C蒸气GAO压消毒),2mL安瓿(或专用细胞冻存管)、吸管、离心管、喷灯、纱布袋(或冻存管架)等。主要操作步骤为:(1)选择处于对数生长期的细胞,在冻存前一天ZuiHAO换。将多个培养瓶中的细胞培养 去掉,用0.25%胰蛋白酶消化。适时去掉胰蛋白酶,加入少量新培养。用吸管吸取培养反复吹打瓶壁上的细胞,使其成为均匀分散的细胞悬。悬浮生产细胞则不要消化处理。然后将细胞收集于离心管中离心(1000r/min,10分钟)。(2)去上清,加入含20%小牛血清的完全培养基,于4℃预冷15分钟后,逐滴加入已无菌的DMSO或甘油,用吸管轻轻吹打使细胞均匀,细胞浓度为3×106~1×107/mL之间。(3)将上述细胞分装于安瓿或专用冷冻塑料管中,安瓿装1~1.5mL在火焰喷灯上封口,封口处要完全封闭,圆滑无勾。冷冻管要将盖子盖紧,并标记HAO细胞名称和冻存日期,同时作HAO登记(日期、细胞种类及代次、冻存支数)。(4)将装HAO细胞的安瓿或冻存管装入沙布袋内;置于容器颈口处存放过夜,次日转入中。采用控制降温速度的方法也可采用下列步骤:先将安瓿置入4℃冰箱中2~3小时,再移至冰箱冷冻室内3~4小时,再吊入容器颈气态部分存放2小时,Zui后沉入中。细胞冻存在中可以长期保存,但为妥善起见,冻存半年后,ZuiHAO取出一只安瓿细胞复苏培养,观察生长情况,然后再继续冻存。
1301 Cells;背景说明:急性T淋巴细胞白血病;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:L cell line细胞、CoCL2细胞、PSN-1细胞
H2122 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:淋巴母细胞;相关产品有:HTR-8/SV-neo细胞、H2023细胞、RMa-bm细胞
CCRF/CEM/0 Cells;背景说明:G.E. Foley 等人建立了类淋巴母细胞细胞株CCRF-CEM。 细胞是1964年11月从一位四岁白人女性急性淋巴细胞白血病患者的外周血白血球衣中得到。此细胞系从香港收集而来。;传代方法:1:2传代。3天内可长满。;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:H9c2(2-1)细胞、HCC-1419细胞、H661细胞
NCI-H1693 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,每周换液2次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:MARC 145细胞、RCK8细胞、Hs888Lu细胞
OCILY7 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:SNU-520细胞、PT-67细胞、Hs839T细胞
B5537SKIN Cells;背景说明:成纤维 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MLTC-1细胞、HT 29细胞、OVCAR-3细胞
Panc-327 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HCEC-12细胞、SN12C细胞、SNU-C2B细胞
HHFK Cells;背景说明:毛囊;角质 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HEMCSS细胞、MDA-MB-134细胞、Michigan Cancer Foundation-10A细胞
HOP62 Cells;背景说明:肺癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:UM-RC-2细胞、SK-OV-3细胞、Ku812F细胞
MLE-12 Cells;背景说明:肺;上皮细胞;SV40转化;FVB/N;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MDA361细胞、Sp 2817细胞、TMK1细胞
NKM1 Cells;背景说明:急性髓系白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:ECC 10细胞、RD-2细胞、766T细胞
Panc04.03 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:NK92细胞、NF639细胞、Rabbit Kidney 13细胞
UMUC1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HLE细胞、HFL-1细胞、H-2029细胞
Medical Research Council cell strain-5 Cells;背景说明:MRC-5细胞系来自14周龄男性胎儿的正常肺组织,该细胞老化前能传代42~46个倍增时间。;传代方法:1:2-1:5传代;每周1-2次。;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:SupT1细胞、G 292 Clone A 141B1细胞、IR983F细胞
Hep-3B Cells;背景说明:肝癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs445细胞、T24细胞、HOS/MNNG细胞
Jurkat-77 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Human fetal lung fibroblast 1细胞、CP-70细胞、686LN-M4e细胞
P3HR1-BL Cells;背景说明:详见相关文献介绍;传代方法:每2-3天换液;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:Pan02细胞、MCA-205细胞、SK-ES-1细胞
Abcam A-549 PBK KO Cells(提供STR鉴定图谱)
ACC-3 Cells(提供STR鉴定图谱)
BayGenomics ES cell line CSH149 Cells(提供STR鉴定图谱)
BayGenomics ES cell line RRU502 Cells(提供STR鉴定图谱)
BC3A Cells(提供STR鉴定图谱)
CHO-K1 Dd-B7 Cells(提供STR鉴定图谱)
DA02565 Cells(提供STR鉴定图谱)
DA05808 Cells(提供STR鉴定图谱)
GM00068 Cells(提供STR鉴定图谱)
H-211 Cells;背景说明:详见相关文献介绍;传代方法:3-4天换液1次。;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:SUM 190细胞、HT1197细胞、CEM C7细胞
OCILY19 Cells;背景说明:弥漫大B淋巴瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:hFOB 1.19细胞、NCIH2029细胞、Walker-256细胞
NCI660 Cells;背景说明:详见相关文献介绍;传代方法:2-3天换液1次。;生长特性:悬浮生长;形态特性:上皮细胞;相关产品有:87 MG细胞、NCI-H23细胞、MV 4;11细胞
Mv 1 Lu Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Sf-21细胞、C4-1细胞、RL-65细胞
Monomac-1 Cells;背景说明:急性单核细胞白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:H2228细胞、Panc 10.05细胞、SUM190细胞
HNE-3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SK ES 01细胞、SK 1细胞、HOS-MNNG细胞
SMMC-7721 Cells;背景说明:用Northernblot方法,未能检测到细胞中1.3kbLFIRE-1/HFREP-1mRNA的表达。;传代方法:1:3传代,2-3天换液一次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:CCD841CoN细胞、NCIH1651细胞、L Wnt-3A细胞
NCIH2291 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:Mouse Colon 38细胞、WB F344细胞、KE37细胞
SW480[SW-480]人结肠癌细胞全年复苏|已有STR图谱
NF639 Cells;背景说明:乳腺癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MKN74细胞、MDAPCa-2b细胞、SJRH 30细胞
OSA Cells;背景说明:详见相关文献介绍;传代方法:1:5-1:10传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:SHIN3细胞、SUDHL-4细胞、OCI-Ly07细胞
SCC90 Cells;背景说明:舌鳞癌细胞;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:COLO.205细胞、U-373MG ATCC细胞、W256细胞
H2227 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代;每周换液2次。;生长特性:该细胞既有悬浮生长,又有贴壁生长;形态特性:上皮细胞;相关产品有:ARPE19细胞、TW01细胞、COLO 741细胞
LWnt3A Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H2195细胞、NCI-H2330细胞、SGC-7901细胞
LWnt3A Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H2195细胞、NCI-H2330细胞、SGC-7901细胞
NCIH1105 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2-3次;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:ST 486细胞、CT26.WT细胞、H-1299细胞
GM3498B-HV1 Cells(提供STR鉴定图谱)
HAP1 PGP (-) 4 Cells(提供STR鉴定图谱)
LICCF Cells;背景说明:肝内胆管癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Emory University-3细胞、DB细胞、EJ1细胞
JAR Cells;背景说明:JAR细胞株来源于胎盘滋养层肿瘤;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:SkChA-1细胞、BL1339细胞、HEL-92.1.7细胞
NCTC clone 1469 Cells;背景说明:1952年建系,源于正常C3H/An小鼠的肝脏组织,表达H-2K抗原,鼠痘病毒阴性。;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:淋巴母细胞;相关产品有:Hs 729T细胞、Centre Antoine Lacassagne-27细胞、WM 239细胞
PLA801-95D Cells;背景说明:这是一株高转移肺癌。;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:SJSA细胞、Panc05.04细胞、MPC 11细胞
RL Cells;背景说明:详见相关文献介绍;传代方法:每周换液2-3次。;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:SCC90细胞、NCIH596细胞、C22细胞
UM-UC-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:FM88细胞、YT细胞、C3H/10T1/2细胞
Hs 729.T Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:H-820细胞、KYSE-50细胞、WM115mel细胞
T98 G Cells;背景说明:详见相关文献介绍;传代方法:按1:3传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:Colon38细胞、MMQ细胞、32D.cl3细胞
HSC-40A Cells(提供STR鉴定图谱)
KOLF2.1J SPG11 PTC PTC/PTC Cells(提供STR鉴定图谱)
MOLT-4/ADR Cells(提供STR鉴定图谱)
NYSCF-10005-1051-1051-Skin-mR-iPSC Cells(提供STR鉴定图谱)
RG-358 Cells(提供STR鉴定图谱)
Ubigene HEK293 BACH1 KO Cells(提供STR鉴定图谱)
WCMC-32 Cells(提供STR鉴定图谱)
HEV0356 Cells(提供STR鉴定图谱)
GP293 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KU 812细胞、CL-11细胞、BMSC/hBMSCs细胞
Hela-mock Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LS-123细胞、HCV 29细胞、Hs 578.Bst细胞
H-727 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:293A细胞、U251细胞、PK-13细胞
AML-EOL-1 Cells;背景说明:急性髓系白血病;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:HSAS1细胞、FAK+/+细胞、NCI-H2009细胞
NCIH1993 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:6传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:3AA细胞、KPL4细胞、HUV-EC-C细胞
NCIH1993 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:6传代;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:3AA细胞、KPL4细胞、HUV-EC-C细胞
NCI-H2228 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,每周2-3次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:HEM细胞、CaES-17细胞、Jiyoye(P-2003)细胞
Hi5 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:JC细胞、SKCO1细胞、CEMx721.174.T2细胞
SNB-19 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:LP1细胞、CCD18细胞、SKOV3细胞
OVCAR 8 Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CCRF-CEM/S细胞、Ball 1细胞、JIMT细胞
NCI-H2081 Cells;背景说明:详见相关文献介绍;传代方法:随细胞的密度而增加;生长特性:悬浮生长;形态特性:聚团悬浮;相关产品有:NCI-H102细胞、Mouse podocyte细胞、Evsa-T细胞
NCIH2122 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:淋巴母细胞;相关产品有:LN-382细胞、OVTOKO细胞、NCIH1581细胞
Clone 166 Cells;背景说明:血管内皮;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CCD-33Co细胞、COLO 320F细胞、SV40 MES 13细胞
Alpha Mouse Liver 12 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:KTC-1细胞、COV-362细胞、EAhy 926细胞
A-10 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Co-115细胞、CF-PAC1细胞、SJ-RH30细胞
SU-ALL-2 Cells(提供STR鉴定图谱)
Jijoye Cells;背景说明:详见相关文献介绍;传代方法:每周2-3次。;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:D283细胞、SRA 01/04细胞、Mo 59J细胞
MCF-10 Attached Cells;背景说明:乳腺;上皮细胞;自发永生;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NCIH929细胞、E6-1细胞、RL952细胞
CT26-clone 25 Cells;背景说明:结肠癌; BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MRCV细胞、Hce8693细胞、SF 539细胞
L-WRN Cells;背景说明:皮下结缔组织;自发永生;雄性;C3H/An;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:LC-1细胞、NCIH1836细胞、CW-2细胞
M059K Cells;背景说明:详见相关文献介绍;传代方法:1:6-1:8传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:NMCG1细胞、C-Lu65细胞、M07e细胞
HEL 92.1.7 Cells;背景说明:详见相关文献介绍;传代方法:每周2-3次。;生长特性:悬浮生长;形态特性:成淋巴细胞;相关产品有:MA104细胞、MSTO-211 H细胞、T-24细胞
M-07e Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KP-2细胞、TGW-nu-1细胞、NCI-H2108细胞
HEL Cells;背景说明:这株淋巴母细胞样细胞株,源自一位30岁白人男性一,患有恶性红细胞白血病,能够自然产生并能诱导球蛋白合成。细胞的EB病毒核抗原阴性,没有表面免疫球蛋白与细胞质免疫球蛋白。HEL细胞表达HLA抗原(HLA-A3,AW32,BW35),β-2小球蛋白,一定比例的细胞还表达Ia抗原。这个细胞株提供了一种用于研究红细胞分化和球蛋白基因表达的模型。它类似于小鼠中的血友病。;传代方法:1:2传代。3天内可长满。;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:OSA细胞、MPP 89细胞、MDAMB175VII细胞
SW480[SW-480]人结肠癌细胞全年复苏|已有STR图谱
H-1650 Cells;背景说明:该细胞是从一名27岁白人男性(10年烟龄)支气管肺泡癌患者的胸腔积液中分离得到的。;传代方法:1:4-1:6传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:IPLB-SF 21细胞、DAKIKI Clone 1细胞、U 266细胞
RAW2647 Cells;背景说明:单核巨噬细胞白血病;雄性;BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:P30/OHK细胞、MDA330细胞、DI TNC-1细胞
WIL2 Secreting Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H-441细胞、BNL-HCC细胞、WIL2-S细胞
Hce8693 Cells;背景说明:盲肠腺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:UPCI-SCC-154细胞、SW-579细胞、U2-OS细胞
DoTc2 4510 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:3传代,每周换液2—3次;生长特性:贴壁生长 ;形态特性:上皮样;相关产品有:RMG1细胞、Liver-02细胞、SHI-1细胞
COLO 16 Cells;背景说明:皮肤鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:PNT1/A细胞、PLA-802细胞、C41细胞
BayGenomics ES cell line RRH037 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XH142 Cells(提供STR鉴定图谱)
ES[MC1R(20):tetControl(13)] Cells(提供STR鉴定图谱)
N-S.7 Cells(提供STR鉴定图谱)
Ubigene MC-38 Cd274 KO Cells(提供STR鉴定图谱)
Mayes Cells(提供STR鉴定图谱)
" "PubMed=924690; DOI=10.1002/ijc.2910200505
Kerbel R.S., Pross H.F., Leibovitz A.
Analysis of established human carcinoma cell lines for lymphoreticular-associated membrane receptors.
Int. J. Cancer 20:673-679(1977)
PubMed=286328; DOI=10.1073/pnas.76.3.1438; PMCID=PMC383267
Herlyn M., Steplewski Z., Herlyn D., Koprowski H.
Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 76:1438-1442(1979)
PubMed=288927; DOI=10.1093/jnci/63.3.635
Leibovitz A., Wright W.C., Pathak S., Siciliano M.J., Daniels W.P., Fogh H., Fogh J.
Detection and analysis of a glucose 6-phosphate dehydrogenase phenotype B cell line contamination.
J. Natl. Cancer Inst. 63:635-645(1979)
PubMed=7459858
Rousset M., Zweibaum A., Fogh J.
Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins.
Cancer Res. 41:1165-1170(1981)
PubMed=7104989; DOI=10.1016/0165-4608(82)90076-0
Chen T.-R., Hay R.J., Macy M.L.
Karyotype consistency in human colorectal carcinoma cell lines established in vitro.
Cancer Genet. Cytogenet. 6:93-117(1982)
PubMed=6582512; DOI=10.1073/pnas.81.2.568; PMCID=PMC344720
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=3349466
Chantret I., Barbat A., Dussaulx E., Brattain M.G., Zweibaum A.
Epithelial polarity, villin expression, and enterocytic differentiation of cultured human colon carcinoma cells: a survey of twenty cell lines.
Cancer Res. 48:1936-1942(1988)
PubMed=1778766; DOI=10.1111/j.1349-7006.1991.tb01816.x; PMCID=PMC5918361
Takeshima E., Hamaguchi M., Watanabe T., Akiyama S., Kataoka M., Ohnishi Y., Xiao H.-Y., Nagai Y., Takagi H.
Aberrant elevation of tyrosine-specific phosphorylation in human gastric cancer cells.
Jpn. J. Cancer Res. 82:1428-1435(1991)
PubMed=1389533; DOI=10.1016/0959-8049(92)90031-V
Lahm H., Petral-Malec D., Yilmaz-Ceyhan A., Fischer J.R., Lorenzoni M., Givel J.-C., Odartchenko N.
Growth stimulation of a human colorectal carcinoma cell line by interleukin-1 and -6 and antagonistic effects of transforming growth factor beta 1.
Eur. J. Cancer 28A:1894-1899(1992)
PubMed=8422623; DOI=10.1002/1097-0142(19930115)71:2<315::aid-cncr2820710208>3.0.CO;2-B
Tibbetts L.M., Doremus C.M., Tzanakakis G.N., Vezeridis M.P.
Liver metastases with 10 human colon carcinoma cell lines in nude mice and association with carcinoembryonic antigen production.
Cancer 71:315-321(1993)
PubMed=7651727
Kastrinakis W.V., Ramchurren N., Rieger K.M., Hess D.T., Loda M., Steele G., Summerhayes I.C.
Increased incidence of p53 mutations is associated with hepatic metastasis in colorectal neoplastic progression.
Oncogene 11:647-652(1995)
PubMed=7824277
Eshleman J.R., Lang E.Z., Bowerfind G.K., Parsons R., Vogelstein B., Willson J.K.V., Veigl M.L., Sedwick W.D., Markowitz S.D.
Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer.
Oncogene 10:33-37(1995)
PubMed=8895552; DOI=10.1002/(SICI)1097-0215(19960927)68:1<126::aid-ijc22>3.0.CO;2-8
Suardet L., Li C., Little J.B.
Radio-induced modulation of transforming growth factor beta1 sensitivity in a p53 wild-type human colorectal-cancer cell line.
Int. J. Cancer 68:126-131(1996)
PubMed=9000147
Cottu P.-H., Muzeau F., Estreicher A., Flejou J.-F., Iggo R.D., Thomas G., Hamelin R.
Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines.
Oncogene 13:2727-2730(1996)
PubMed=9000572
Hoang J.-M., Cottu P.-H., Thuille B., Salmon R.J., Thomas G., Hamelin R.
BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines.
Cancer Res. 57:300-303(1997)
PubMed=9290701; DOI=10.1002/(SICI)1098-2744(199708)19:4<243::aid-mc5>3.0.CO;2-D
Jia L.-Q., Osada M., Ishioka C., Gamo M., Ikawa S., Suzuki T., Shimodaira H., Niitani T., Kudo T., Akiyama M., Kimura N., Matsuo M., Mizusawa H., Tanaka N., Koyama H., Namba M., Kanamaru R., Kuroki T.
Screening the p53 status of human cell lines using a yeast functional assay.
Mol. Carcinog. 19:243-253(1997)
PubMed=9294210; DOI=10.1073/pnas.94.19.10330; PMCID=PMC23362
Ilyas M., Tomlinson I.P.M., Rowan A.J., Pignatelli M., Bodmer W.F.
Beta-catenin mutations in cell lines established from human colorectal cancers.
Proc. Natl. Acad. Sci. U.S.A. 94:10330-10334(1997)
PubMed=9715273; DOI=10.1038/sj.onc.1201986
Eshleman J.R., Casey G., Kochera M.E., Sedwick W.D., Swinler S.E., Veigl M.L., Willson J.K.V., Schwartz S., Markowitz S.D.
Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53.
Oncogene 17:719-725(1998)
PubMed=10674020; DOI=10.1016/S0959-8049(99)00206-3
Ku J.-L., Yoon K.-A., Kim D.-Y., Park J.-G.
Mutations in hMSH6 alone are not sufficient to cause the microsatellite instability in colorectal cancer cell lines.
Eur. J. Cancer 35:1724-1729(1999)
PubMed=10612807; DOI=10.1002/(SICI)1098-2264(200002)27:2<183::aid-gcc10>3.0.CO;2-P; PMCID=PMC4721570
Ghadimi B.M., Sackett D.L., Difilippantonio M.J., Schrock E., Neumann T., Jauho A., Auer G., Ried T.
Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations.
Genes Chromosomes Cancer 27:183-190(2000)
PubMed=10737795; DOI=10.1073/pnas.97.7.3352; PMCID=PMC16243
Rowan A.J., Lamlum H., Ilyas M., Wheeler J.M.D., Straub J., Papadopoulou A., Bicknell D.C., Bodmer W.F., Tomlinson I.P.M.
APC mutations in sporadic colorectal tumors: a mutational 'hotspot' and interdependence of the 'two hits'.
Proc. Natl. Acad. Sci. U.S.A. 97:3352-3357(2000)
PubMed=10773689; DOI=10.1159/000015508
Melcher R., Steinlein C., Feichtinger W., Muller C.R., Menzel T., Luehrs H., Scheppach W., Schmid M.
Spectral karyotyping of the human colon cancer cell lines SW480 and SW620.
Cytogenet. Cell Genet. 88:145-152(2000)
PubMed=11113861; DOI=10.1002/1096-9896(2000)9999:9999<::aid-path775>3.0.CO;2-K
Hewitt R.E., McMarlin A., Kleiner D.E., Wersto R.P., Martin P., Tsokos M.G., Stamp G.W.H., Stetler-Stevenson W.G.
Validation of a model of colon cancer progression.
J. Pathol. 192:446-454(2000)
PubMed=11226274; DOI=10.1073/pnas.041603298; PMCID=PMC30173
Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D.C., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.W.
Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement.
Proc. Natl. Acad. Sci. U.S.A. 98:2538-2543(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11526487; DOI=10.1038/sj.onc.1204611
Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P.-H., Hamelin R.
Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines.
Oncogene 20:5025-5032(2001)
PubMed=11668190; DOI=10.1177/002215540104901105
Quentmeier H., Osborn M., Reinhardt J., Zaborski M., Drexler H.G.
Immunocytochemical analysis of cell lines derived from solid tumors.
J. Histochem. Cytochem. 49:1369-1378(2001)
PubMed=16418264; DOI=10.1073/pnas.0510146103; PMCID=PMC1327731
Liu Y., Bodmer W.F.
Analysis of p53 mutations and their expression in 56 colorectal cancer cell lines.
Proc. Natl. Acad. Sci. U.S.A. 103:976-981(2006)
PubMed=16854228; DOI=10.1186/1476-4598-5-29; PMCID=PMC1550420
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=19927377; DOI=10.1002/gcc.20730; PMCID=PMC2818350
Knutsen T., Padilla-Nash H.M., Wangsa D., Barenboim-Stapleton L., Camps J., McNeil N.E., Difilippantonio M.J., Ried T.
Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines.
Genes Chromosomes Cancer 49:204-223(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192; PMCID=PMC2943514
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780; PMCID=PMC2920028
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=20831567; DOI=10.1111/j.1582-4934.2010.01170.x; PMCID=PMC3918049
Ma Y.-L., Zhang P., Wang F., Moyer M.P., Yang J.-J., Liu Z.-H., Peng J.-Y., Chen H.-Q., Zhou Y.-K., Liu W.-J., Qin H.-L.
Human embryonic stem cells and metastatic colorectal cancer cells shared the common endogenous human microRNA-26b.
J. Cell. Mol. Med. 15:1941-1954(2011)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66; PMCID=PMC3543849
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=23932154; DOI=10.1016/j.radonc.2013.06.032
Salendo J., Spitzner M., Kramer F., Zhang X., Jo P., Wolff H.A., Kitz J., Kaulfuss S., Beissbarth T., belstein M., Ghadimi M., Grade M., Gaedcke J.
Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g.
Radiother. Oncol. 108:451-457(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35; PMCID=PMC3816225
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=24840470; DOI=10.1016/j.jprot.2014.05.002
Chik J.H.L., Zhou J., Moh E.S.X., Christopherson R., Clarke S.J., Molloy M.P., Packer N.H.
Comprehensive glycomics comparison between colon cancer cell cultures and tumours: implications for biomarker studies.
J. Proteomics 108:146-162(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25841592; DOI=10.1016/j.jprot.2015.03.019
Piersma S.R., Knol J.C., de Reus I., Labots M., Sampadi B.K., Pham T.V., Ishihama Y., Verheul H.M.W., Jimenez C.R.
Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines.
J. Proteomics 127:247-258(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235; PMCID=PMC4762531
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27987026; DOI=10.1007/s00216-016-0125-5; PMCID=PMC5303640
Schunter A.J., Yue X.-S., Hummon A.B.
Phosphoproteomics of colon cancer metastasis: comparative mass spectrometric analysis of the isogenic primary and metastatic cell lines SW480 and SW620.
Anal. Bioanal. Chem. 409:1749-1763(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y; PMCID=PMC5498998
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=29101300; DOI=10.15252/msb.20177701; PMCID=PMC5731344
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D.J., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29131639; DOI=10.1021/acs.jproteome.7b00548
Torres S., Garcia-Palmero I., Marin-Vicente C., Bartolome R.A., Calvino E., Fernandez-Acenero M.J., Casal J.I.
Proteomic characterization of transcription and splicing factors associated with a metastatic phenotype in colorectal cancer.
J. Proteome Res. 17:252-264(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31059103; DOI=10.3892/or.2019.7146
Wang Y.-Y., Zhou L., Qing Q., Li Y.-F., Li L.-X., Dong X.-Y., Xiao B.
Gene expression profile of cancer stem-like cells in the SW480 colon adenocarcinoma cell line.
Oncol. Rep. 42:386-398(2019)"
![SW480 [SW-480] (人结肠癌细胞)(L15)](https://img.chemicalbook.com/SupplyImg/2022-10-26/Large/202210261007361367032.jpg)
![SW480[SW-480]细胞](https://img.chemicalbook.com/SupplyImg/2023-09-28/Large/202309281103335738061.png)

![SW480[SW-480]人结肠癌复苏细胞|送STR报告](https://img.chemicalbook.com/SupplyImg1/2025-02-16/Large/202502160837329976570.jpg)
![SW480[SW-480]细胞系|人结直肠腺癌细胞](https://img.chemicalbook.com/SupplyImg/2019-10-07/Large/201910071539463926848.jpg)
![SW480[SW-480]细胞(结肠癌细胞STR鉴定正确)LM8C0227](https://img.chemicalbook.com/SupplyImg2/2025-12-04/Large/202512041059288498139.jpg)
YB-71605HC](https://img.chemicalbook.com/SupplyImg1/2025-08-06/Large/202508061354042928244.jpg)
![SW480[SW-480]人结肠癌细胞库|培养基|带STR图谱](https://img.chemicalbook.com/SupplyImg1/2025-02-01/Large/202502011936419894429.jpg)
![SW480 [SW-480] 人结肠癌细胞](https://img.chemicalbook.com/SupplyImg1/2025-08-27/Large/202508271041054252712.png)

![SW480[SW-480]人结肠癌细胞全年复苏|已有STR图谱](/SupplyImg1/2025-02-22/202502220320211985850.jpg)
