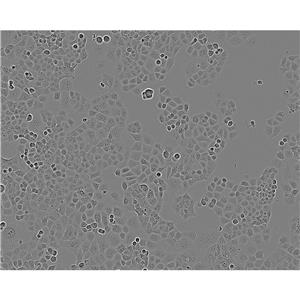
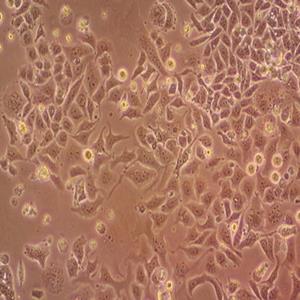
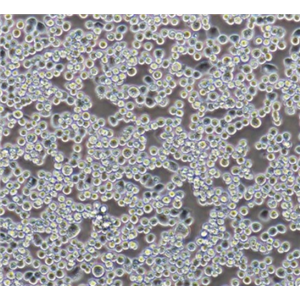
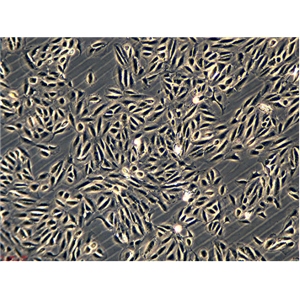
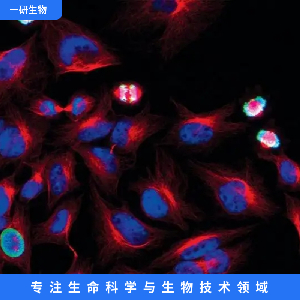
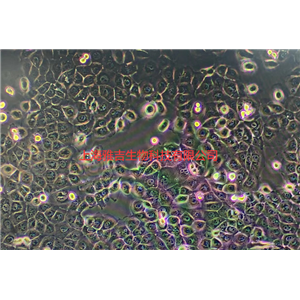
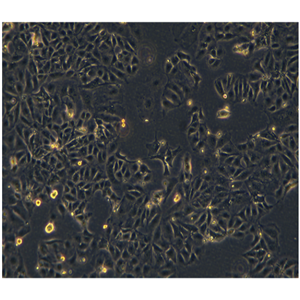
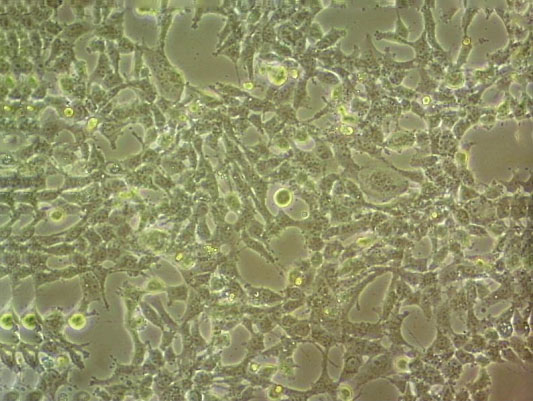
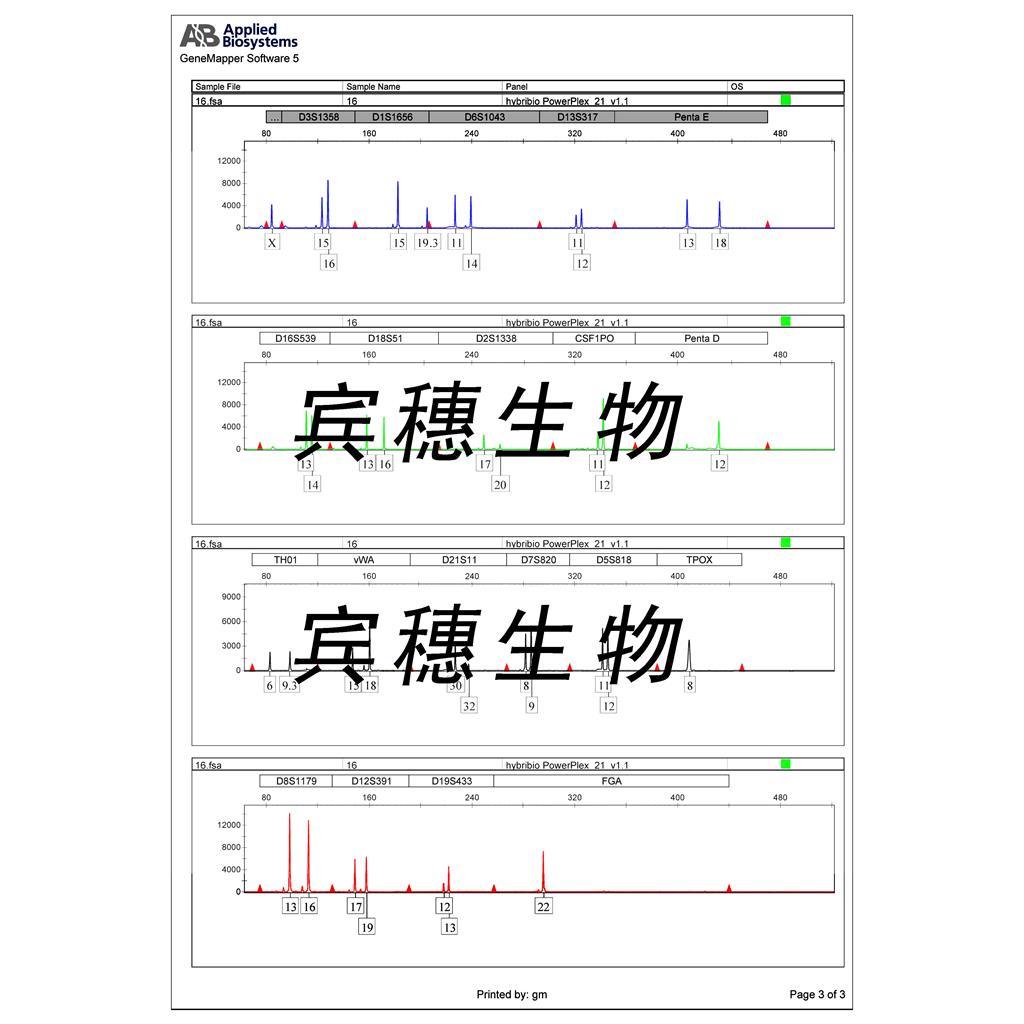
"SW620/LUC人结肠癌细胞代次低|培养基|送STR图谱<荧光素酶标记>
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
【细胞培养经验分享】启蒙老师的重要性:一般进实验室都有师兄师姐带着做,他们就是你做细胞的启蒙老师。他们的操作手法、细节、理论讲解就成了你操作的准则,如营养液、细胞瓶的摆放位置、灭菌处理程序、开盖手法、细胞吹打手法等等。要学会他们的正确操作,在第一次的时候就要重视。像养孩子一样养细胞,细胞有时真的很脆弱,最好每天都去看看它,以防止出现培养箱缺水、缺二氧化碳、停电、温度不够等异常现象,也好及时解决这些意外,避免重复实验带来的更大痛苦。好细胞要及时保种:细胞要分批传代,这样即使有一批出了问题,还有一批备用的。像后者一般人可能不容易做到。但这是我血的教训,有一次细胞污染了,全军覆没。当时可后悔没有保种。细胞跟人一样,不同的细胞,培养特性是不一样的。培养过程中要细细体会,不同细胞系使用不同的培养基和血清。
换液周期:每周2-3次
U266 Bl Cells;背景说明:详见相关文献介绍;传代方法:1:3传代,2-3天传一代;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:H1651细胞、TM4细胞、SU.86.86细胞
SPCA-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Hs-688A-T细胞、BC-020细胞、CAL62细胞
Hs940-T Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:NCI-H2228细胞、SNU-886细胞、DMS 79细胞
SW620/LUC人结肠癌细胞代次低|培养基|送STR图谱<荧光素酶标记>
背景信息:详见相关文献介绍
┈订┈购(技术服务)┈热┈线:1┈3┈6┈4┈1┈9┈3┈0┈7┈9┈1【微信同号】┈Q┈Q:3┈1┈8┈0┈8┈0┈7┈3┈2┈4;
贴壁细胞的传代培养,详细步骤如下:首先倒掉培养基,在这一步骤可以收集一些细胞上清做支原体检测;加入胰蛋白酶,一般T25是加2mL,盖好瓶盖,摇晃T25培养瓶,使胰蛋白酶均匀覆盖在细胞表面,放入培养箱2-3min,期间可在显微镜下观察,看到大部分细胞变圆,即可放入超净台,加入2倍的完全培养基,这里就是加4mL培养基,终止消化;将含有胰蛋白酶,细胞和培养基一起转移到离心管中,1000rpm/3min离心,去掉上清;新鲜的完全培养基重悬,根据细胞的生长特性和后续的实验需求进行传代,比如我养的Hepa1-6就长的比较快,不是着急用的话,我就会按1E6个细胞/T75培养瓶进行传代;但如果后两天要用,就会适当多传一点;还可通过显微镜计数后,直接用于细胞铺板,继续后续的实验。
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
SW620/LUC人结肠癌细胞代次低|培养基|送STR图谱<荧光素酶标记>
物种来源:人源、鼠源等其它物种来源
DHL-6 Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:6传代,3—4天换液1次;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:MLMA细胞、H719细胞、A549/DDP细胞
OACP4C Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:B-cell Acute Lymphoblastic Leukemia-1细胞、EOC 20细胞、NB-4细胞
Kit225/K6 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:J111细胞、GM00215细胞、OCI-AML4细胞
MiaPaCa-2 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:TW-039细胞、FET细胞、Potorous tridactylus Kidney 1细胞
┈订┈购(技术服务)┈热┈线:1┈3┈6┈4┈1┈9┈3┈0┈7┈9┈1【微信同号】┈Q┈Q:3┈1┈8┈0┈8┈0┈7┈3┈2┈4;
形态特性:上皮细胞样
细胞冻存复苏材料与方法步骤:常用的细胞冷冻贮存器为贮存器,规格有35L和50L两种。使用时要注意以下几点:(1)一般两周需充一次,至少一个月充一次。温度达-196℃,使用时注意勿让溅到皮肤上,以免引起冻伤。(2)容器为双层结构,中间为真空层,瓶口有双层焊接处,应防止焊接部裂开。(3)在装入时,要注意缓慢小心,并用厚纸卷筒或制漏斗作引导,使直达瓶底,如有专用灌注装置则更HAO。若为初次使用,加时更要缓慢,以免温度骤降而使容器损坏。细胞冻存时常备的材料有:0.25%胰蛋白酶,含10%~20%的血清培养,DMSO(分析纯)或无色新鲜甘油(121°C蒸气GAO压消毒),2mL安瓿(或专用细胞冻存管)、吸管、离心管、喷灯、纱布袋(或冻存管架)等。主要操作步骤为:(1)选择处于对数生长期的细胞,在冻存前一天ZuiHAO换。将多个培养瓶中的细胞培养 去掉,用0.25%胰蛋白酶消化。适时去掉胰蛋白酶,加入少量新培养。用吸管吸取培养反复吹打瓶壁上的细胞,使其成为均匀分散的细胞悬。悬浮生产细胞则不要消化处理。然后将细胞收集于离心管中离心(1000r/min,10分钟)。(2)去上清,加入含20%小牛血清的完全培养基,于4℃预冷15分钟后,逐滴加入已无菌的DMSO或甘油,用吸管轻轻吹打使细胞均匀,细胞浓度为3×106~1×107/mL之间。(3)将上述细胞分装于安瓿或专用冷冻塑料管中,安瓿装1~1.5mL在火焰喷灯上封口,封口处要完全封闭,圆滑无勾。冷冻管要将盖子盖紧,并标记HAO细胞名称和冻存日期,同时作HAO登记(日期、细胞种类及代次、冻存支数)。(4)将装HAO细胞的安瓿或冻存管装入沙布袋内;置于容器颈口处存放过夜,次日转入中。采用控制降温速度的方法也可采用下列步骤:先将安瓿置入4℃冰箱中2~3小时,再移至冰箱冷冻室内3~4小时,再吊入容器颈气态部分存放2小时,Zui后沉入中。细胞冻存在中可以长期保存,但为妥善起见,冻存半年后,ZuiHAO取出一只安瓿细胞复苏培养,观察生长情况,然后再继续冻存。
HEK293-EBNA Cells;背景说明:详见相关文献介绍;传代方法:1:4-1:10传代;每周2次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:Panc 2.03细胞、Hs746T细胞、U-937细胞
WEHI 3 Cells;背景说明:白血病;BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCI-H292细胞、Hs27F细胞、Clone 15 HL-60细胞
H-2023 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Kelly细胞、NCI-H102细胞、Reh细胞
TE-7 Cells;背景说明:食管鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Human Intestinal Epithelial Cell-6细胞、MGH-UI细胞、H2291细胞
NCI-H520 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;2-3天换液1次;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Hs888T细胞、DC2.4细胞、HIEC-6细胞
HMEC-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:LICCF细胞、5637细胞、APRE-19细胞
NUGC4 Cells;背景说明:胃癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:半贴壁;形态特性:详见产品说明书;相关产品有:HCEpiC细胞、IGROV-1细胞、TGW-nu-1细胞
HEL 299 Cells;背景说明:红白细胞白血病;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:H2085细胞、MM.1S细胞、L-5178-Y细胞
McA-RH7777 Cells;背景说明:肝癌;雌性;Buffalo;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-1373细胞、SUM-149细胞、SMC-1细胞
NCI-H727 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:T-ALL 1细胞、NCI-H1993细胞、KG-1细胞
M619 Cells;背景说明:脉络膜黑色素瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H498细胞、PC-10细胞、UM-RC-2细胞
SK-MEL2 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:多边形的;相关产品有:COLO824细胞、NCIH2286细胞、NBL-2细胞
HRGEC Cells;背景说明:肾小球;内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:OCI-LY-18细胞、HTR8svn细胞、RERF-LCMS细胞
OUMS-23 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:210RCY3-Ag1.2.3细胞、SF 763细胞、COLO206F细胞
RDES-1 Cells;背景说明:详见相关文献介绍;传代方法:1:8传代:每周换液2-3次;生长特性:贴壁和悬浮混合;形态特性:上皮样;相关产品有:HuT 78细胞、U-373-MG细胞、C-Lu-65细胞
108CC15 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:OAW 42细胞、BHP10-3细胞、RT 112细胞
Cor L51 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NU-GC-3细胞、8305C细胞、MeSoTheliOma-211H细胞
Abcam HCT 116 GATA2 KO Cells(提供STR鉴定图谱)
AG06846 Cells(提供STR鉴定图谱)
BayGenomics ES cell line CSI690 Cells(提供STR鉴定图谱)
BayGenomics ES cell line STA078 Cells(提供STR鉴定图谱)
BIONi010-C-29 Cells(提供STR鉴定图谱)
CIPGC Cells(提供STR鉴定图谱)
DA03255 Cells(提供STR鉴定图谱)
DD2173 Cells(提供STR鉴定图谱)
GM02078 Cells(提供STR鉴定图谱)
HSCT6 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:LSECs细胞、BI-Mel细胞、SVGp12细胞
SW620/LUC人结肠癌细胞代次低|培养基|送STR图谱<荧光素酶标记>
SU86_86 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:HFL-I细胞、Melan-a细胞、L-Wnt3A细胞
A9 Cells;背景说明:皮下结缔组织;自发永生;雄性;C3H/An;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NCIH1688细胞、H-1573细胞、A-10细胞
RPTEC/TERT1 Cells;背景说明:肾;近端小管上皮;HGNC-TERT转化;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:PTK-2细胞、H1819细胞、NT2细胞
NFS 60 Cells;背景说明:详见相关文献介绍;传代方法:1:3传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:CHO细胞、NCI-SNU-761细胞、Malme3M细胞
3T3L1 Cells;背景说明:3T3-L1是从3T3细胞(Swissalbino)中经克隆分离得到的连续传代的亚系。该细胞从快速分裂到汇合和接触性抑制状态经历了前脂肪细胞到脂肪样细胞的转变。该细胞鼠痘病毒阴性;可产生甘油三酯,高浓度血清可增强细胞内脂肪堆积。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:CATHa细胞、RKOAS451细胞、HCC-2185细胞
42.9E5.2 Cells(提供STR鉴定图谱)
ATDC-5 Cells;背景说明:详见相关文献介绍;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:KMST-6细胞、OVCAR-3细胞、MBMEC细胞
P30-OHK Cells;背景说明:详见相关文献介绍;传代方法:10^5 cells/60mm dish;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:MDA468细胞、KU812-F细胞、H441细胞
MDAPCa2b Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:TE3细胞、JM1细胞、Lieming Xu-2细胞
Pa18C Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HS-766T细胞、BSC40细胞、NCI-441细胞
HL1 Cells;背景说明:心肌;SV40转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:EM-3细胞、KU-812F细胞、Human Foreskin Fibroblast细胞
NCIH2135 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MES-SA-Dx5细胞、OE-21细胞、MD Anderson-Metastatic Breast-468细胞
J82 COT Cells;背景说明:电子显微镜下未观察到桥粒但观察到数目不同的粗面内质网和突出微丝。 含ras (H-ras)癌基因。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:TE-1细胞、Namalva细胞、NCIH196细胞
WI-38 Cells;背景说明:LeonardHayflick建系;有限传代细胞系;寿命为50±10代(倍增时间24h);来自妊娠3个月的正常胚胎肺组织。该细胞系是第一个用于人制备的人二倍体细胞;培养基中添加TNFα可以加快细胞生长。;传代方法:1:2-1:4传代;2-3天换液1次;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:RD细胞、LMTK-细胞、OCI-AML3细胞
GM17731 Cells(提供STR鉴定图谱)
HAP1 HSPB1 (-) 3 Cells(提供STR鉴定图谱)
RPMI8402 Cells;背景说明:急性T淋巴细胞白血病;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:C-33 A细胞、MDCC MSB1细胞、Rat 1细胞
Psi2 DAP Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCIH128细胞、A9(Hamprecht)细胞、ML2细胞
SUD6 Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:6传代,3—4天换液1次;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:NCI-H1623细胞、RCS细胞、Walker-256-TC细胞
HTh-74 Cells;背景说明:未分化甲状腺癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Mel-MeWo细胞、HSC-I细胞、YD-15细胞
MPP-89 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Adeno-293细胞、COLO 738细胞、IBRS2细胞
TE-13 Cells;背景说明:食管鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Jurkat clone A3细胞、HOS (TE85)细胞、NRK细胞
mousepodocyte Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:HH [Human lymphoma]细胞、Menschliche Und Tierische Zellkulture-3细胞、MCA-38细胞
T/G HA VSMC Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:10T1/2细胞、AsPC-1细胞、HARAB细胞
HPS0172 Cells(提供STR鉴定图谱)
JHU173i Cells(提供STR鉴定图谱)
MDA-MB-231 25PACR Cells(提供STR鉴定图谱)
ND34116 Cells(提供STR鉴定图谱)
PR01241 Cells(提供STR鉴定图谱)
Ubigene HEK293 HCK KO Cells(提供STR鉴定图谱)
WG1534 Cells(提供STR鉴定图谱)
HBEC1-KT Cells(提供STR鉴定图谱)
Detroit-562 Cells;背景说明:器官:咽头 疾病:癌 取材转移灶:胸水;传代方法:1:2-1:4传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:T98细胞、H-35 Reuber细胞、QGP-1细胞
NCIH2126 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代,每周2-3次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:GM05862细胞、H1954细胞、HS-766T细胞
H-157 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:U266 B1细胞、NCI-H1876细胞、Panc10.05细胞
SF-539 BT Cells;背景说明:胶质瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SCL1细胞、Liver-02细胞、RMC细胞
THLE-3 Cells;背景说明:详见相关文献介绍;传代方法:消化3-5分钟。1:2。3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Human podocyte细胞、SKMES细胞、95D细胞
THLE-3 Cells;背景说明:详见相关文献介绍;传代方法:消化3-5分钟。1:2。3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Human podocyte细胞、SKMES细胞、95D细胞
GM03671C Cells;背景说明:G.E. Foley 等人建立了类淋巴母细胞细胞株CCRF-CEM。 细胞是1964年11月从一位四岁白人女性急性淋巴细胞白血病患者的外周血白血球衣中得到。此细胞系从香港收集而来。;传代方法:1:2传代。3天内可长满。;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:JTC-28细胞、MCF7ADR细胞、U-118 MG细胞
THLE-3 Cells;背景说明:详见相关文献介绍;传代方法:消化3-5分钟。1:2。3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Human podocyte细胞、SKMES细胞、95D细胞
NCI-H2195 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Menschliche Und Tierische Zellkulture-3细胞、HEK 293 EBNA细胞、VMCUB1细胞
H-647 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;每周换液2次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:Kit225/K6细胞、P3X63 AG 8.653细胞、NRK49F细胞
THLE-3 Cells;背景说明:详见相关文献介绍;传代方法:消化3-5分钟。1:2。3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Human podocyte细胞、SKMES细胞、95D细胞
THLE-2 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:TALL-1 [Human adult T-ALL]细胞、EBC-1细胞、NPC-TW039细胞
HRMC Cells;背景说明:肾;系膜 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Institute for Medical Research-90细胞、3T3-Swiss细胞、Hs852T细胞
BEL-7402 Cells;背景说明:BEL-7402细胞株是1974年从临床肝癌手术标本中建立的。;传代方法:消化3-5分钟,1:2,3天内可长满;生长特性:贴壁生长;形态特性:上皮样;相关产品有:5-8F细胞、P-36细胞、Epithelioma Papulosum Cyprini细胞
OCUM-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:MDA-MB-157细胞、NCI-H1666细胞、HEK (AD293)细胞
SKOV-3/Luc Cells(提供STR鉴定图谱)
NPA 87 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:L-6TG细胞、32Dcl3细胞、Swiss 3T3细胞
NCI-H209 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代;每周换液2次。;生长特性:悬浮生长,有少数细胞疏松贴壁;形态特性:上皮样;相关产品有:RA.1细胞、H1693细胞、KBM-7细胞
HCE Cells;背景说明:角膜上皮细胞;Ad-SV40转化;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HEC-1-A细胞、SMA560细胞、LUDLU-1细胞
H358 Cells;背景说明:1981年从一位开始化疗之前的患者的肿瘤组织中分离建株。超微结构研究表明细胞质中有Clara细胞的特征结构细胞表达主要的肺表面结合蛋白SP-A的蛋白和RNA。不表达SP-B和SP-C。他们在软琼脂中的克隆形成效率为0.83%。;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:P3/NS1/1-Ag4.1细胞、NK-92 transfected with MFG-hIL2细胞、ACC-2细胞
GM0637 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SUM149细胞、TOV112D细胞、Leukemic 1210细胞
B16BL-6 Cells;背景说明:黑色素瘤;雄性;C57BL/6;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MMQ细胞、Ca 9-22细胞、H1755细胞
SW620/LUC人结肠癌细胞代次低|培养基|送STR图谱<荧光素酶标记>
CORL88 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:H-1819细胞、Vx2细胞、MESSA/Dx5细胞
BT-549 Cells;背景说明:该细胞1978年由W.G.Coutinho和E.Y.Lasfargues建系,源自一位72岁患有乳腺导管癌的白人女性,来源组织包括乳头及浸润导管。该细胞形态包括上皮样细胞及多核巨细胞,可分泌一种粘性物质。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:H719细胞、Karpas-422细胞、Dami细胞
SW 1353 Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs 729.T细胞、WM266-4细胞、alphaTC clone 6细胞
MFD-1 Cells;背景说明:详见相关文献介绍;传代方法:维持细胞浓度在3×104-5×105 cells/ml;2-3天换液1次。;生长特性:悬浮生长;形态特性:淋巴母细胞样 ;相关产品有:LAPC-4细胞、H-128细胞、HCC0038细胞
HUT-125 Cells;背景说明:腺鳞状肺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HEL-92_1_7细胞、HCC0366细胞、NCI-H841细胞
HS 683T Cells;背景说明:该细胞源自76岁白人男性的左颞叶侧胶质瘤组织,有微绒毛,无桥粒。 ;传代方法:1:4传代,每周换液2次;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:NCI-H522细胞、MCF-12F细胞、SKRC-52细胞
HCC-1395 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代,每周换液2—3次;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Calu3细胞、CTLL(2)细胞、MHCC97H细胞
HIT T15 Cells;背景说明:胰岛β细胞;SV40转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:GP2d细胞、NCIH2347细胞、HCC-1438细胞
BayGenomics ES cell line RRF280 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XG499 Cells(提供STR鉴定图谱)
EC-Trpm5tm1Dgen Cells(提供STR鉴定图谱)
MQ49 Cells(提供STR鉴定图谱)
TPC417 Cells(提供STR鉴定图谱)
M4Be T9P76 Cells(提供STR鉴定图谱)
" "PubMed=924690; DOI=10.1002/ijc.2910200505
Kerbel R.S., Pross H.F., Leibovitz A.
Analysis of established human carcinoma cell lines for lymphoreticular-associated membrane receptors.
Int. J. Cancer 20:673-679(1977)
PubMed=286328; DOI=10.1073/pnas.76.3.1438; PMCID=PMC383267
Herlyn M., Steplewski Z., Herlyn D., Koprowski H.
Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 76:1438-1442(1979)
PubMed=288927; DOI=10.1093/jnci/63.3.635
Leibovitz A., Wright W.C., Pathak S., Siciliano M.J., Daniels W.P., Fogh H., Fogh J.
Detection and analysis of a glucose 6-phosphate dehydrogenase phenotype B cell line contamination.
J. Natl. Cancer Inst. 63:635-645(1979)
PubMed=6582512; DOI=10.1073/pnas.81.2.568; PMCID=PMC344720
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=7972006; DOI=10.1073/pnas.91.23.11045; PMCID=PMC45163
Okamoto A., Demetrick D.J., Spillare E.A., Hagiwara K., Hussain S.P., Bennett W.P., Forrester K., Gerwin B.I., Serrano M., Beach D.H., Harris C.C.
Mutations and altered expression of p16INK4 in human cancer.
Proc. Natl. Acad. Sci. U.S.A. 91:11045-11049(1994)
PubMed=9023415; DOI=10.1006/cimm.1996.1062
Seki N., Hoshino T., Kikuchi M., Hayashi A., Itoh K.
HLA-A locus-restricted and tumor-specific CTLs in tumor-infiltrating lymphocytes of patients with non-small cell lung cancer.
Cell. Immunol. 175:101-110(1997)
PubMed=9178645; DOI=10.1006/cimm.1997.1108
Nakao M., Sata M., Saitsu H., Yutani S., Kawamoto M., Kojiro M., Itoh K.
CD4+ hepatic cancer-specific cytotoxic T lymphocytes in patients with hepatocellular carcinoma.
Cell. Immunol. 177:176-181(1997)
PubMed=9290701; DOI=10.1002/(SICI)1098-2744(199708)19:4<243::AID-MC5>3.0.CO;2-D
Jia L.-Q., Osada M., Ishioka C., Gamo M., Ikawa S., Suzuki T., Shimodaira H., Niitani T., Kudo T., Akiyama M., Kimura N., Matsuo M., Mizusawa H., Tanaka N., Koyama H., Namba M., Kanamaru R., Kuroki T.
Screening the p53 status of human cell lines using a yeast functional assay.
Mol. Carcinog. 19:243-253(1997)
PubMed=9294210; DOI=10.1073/pnas.94.19.10330; PMCID=PMC23362
Ilyas M., Tomlinson I.P.M., Rowan A.J., Pignatelli M., Bodmer W.F.
Beta-catenin mutations in cell lines established from human colorectal cancers.
Proc. Natl. Acad. Sci. U.S.A. 94:10330-10334(1997)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10773689; DOI=10.1159/000015508
Melcher R., Steinlein C., Feichtinger W., Muller C.R., Menzel T., Luehrs H., Scheppach W., Schmid M.
Spectral karyotyping of the human colon cancer cell lines SW480 and SW620.
Cytogenet. Cell Genet. 88:145-152(2000)
PubMed=11113861; DOI=10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K
Hewitt R.E., McMarlin A., Kleiner D.E., Wersto R.P., Martin P., Tsokos M.G., Stamp G.W.H., Stetler-Stevenson W.G.
Validation of a model of colon cancer progression.
J. Pathol. 192:446-454(2000)
PubMed=11226274; DOI=10.1073/pnas.041603298; PMCID=PMC30173
Abdel-Rahman W.M., Katsura K., Rens W., Gorman P.A., Sheer D., Bicknell D.C., Bodmer W.F., Arends M.J., Wyllie A.H., Edwards P.A.W.
Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement.
Proc. Natl. Acad. Sci. U.S.A. 98:2538-2543(2001)
PubMed=11414198; DOI=10.1007/s004320000207
Lahm H., Andre S., Hoeflich A., Fischer J.R., Sordat B., Kaltner H., Wolf E., Gabius H.-J.
Comprehensive galectin fingerprinting in a panel of 61 human tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures.
J. Cancer Res. Clin. Oncol. 127:375-386(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12107847; DOI=10.1038/sj.bjc.6600459; PMCID=PMC2376111
Sekhon J., Pereira P.A., Sabbaghian N., Schievella A.R., Rozen R.
Antisense inhibition of methylenetetrahydrofolate reductase reduces survival of methionine-dependent tumour lines.
Br. J. Cancer 87:225-230(2002)
PubMed=15748285; DOI=10.1186/1479-5876-3-11; PMCID=PMC555742
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. 3rd, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=16854228; DOI=10.1186/1476-4598-5-29; PMCID=PMC1550420
Bandres Elizalde E.M., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., Garcia-Foncillas J.
Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues.
Mol. Cancer 5:29.1-29.10(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=18258742; DOI=10.1073/pnas.0712176105; PMCID=PMC2268141
Emaduddin M., Bicknell D.C., Bodmer W.F., Feller S.M.
Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells.
Proc. Natl. Acad. Sci. U.S.A. 105:2358-2362(2008)
PubMed=19188929; DOI=10.1038/tpj.2008.20
Guo J., Anderson M.G., Tapang P., Palma J.P., Rodriguez L.E., Niquette A.L., Li J.-L., Bouska J.J., Wang G., Semizarov D., Albert D.H., Donawho C.K., Glaser K.B., Shah O.J.
Identification of genes that confer tumor cell resistance to the aurora B kinase inhibitor, AZD1152.
Pharmacogenomics J. 9:90-102(2009)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921; PMCID=PMC4020356
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=19927377; DOI=10.1002/gcc.20730; PMCID=PMC2818350
Knutsen T., Padilla-Nash H.M., Wangsa D., Barenboim-Stapleton L., Camps J., McNeil N.E., Difilippantonio M.J., Ried T.
Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines.
Genes Chromosomes Cancer 49:204-223(2010)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20570890; DOI=10.1158/0008-5472.CAN-10-0192; PMCID=PMC2943514
Janakiraman M., Vakiani E., Zeng Z.-S., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C.M., Persaud Y., Levine D.A., Fagin J.A., Jhanwar S.C., Mariadason J.M., Lash A., Ladanyi M., Saltz L.B., Heguy A., Paty P.B., Solit D.B.
Genomic and biological characterization of exon 4 KRAS mutations in human cancer.
Cancer Res. 70:5901-5911(2010)
PubMed=20606684; DOI=10.1038/sj.bjc.6605780; PMCID=PMC2920028
Bracht K., Nicholls A.M., Liu Y., Bodmer W.F.
5-fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency.
Br. J. Cancer 103:340-346(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21912889; DOI=10.1007/s10637-011-9744-z
Sutherland H.S., Hwang I.Y., Marshall E.S., Lindsay B.S., Denny W.A., Gilchrist C., Joseph W.R., Greenhalgh D., Richardson E., Kestell P., Ding A., Baguley B.C.
Therapeutic reactivation of mutant p53 protein by quinazoline derivatives.
Invest. New Drugs 30:2035-2045(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096; PMCID=PMC3285665
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595; PMCID=PMC3526189
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=23272949; DOI=10.1186/1755-8794-5-66; PMCID=PMC3543849
Schlicker A., Beran G., Chresta C.M., McWalter G., Pritchard A., Weston S., Runswick S., Davenport S., Heathcote K., Castro D.A., Orphanides G., French T., Wessels L.F.A.
Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines.
BMC Med. Genomics 5:66.1-66.15(2012)
PubMed=23631600; DOI=10.1021/pr400260h
Loftus N.J., Lai L., Wilkinson R.W., Odedra R., Wilson I.D., Barnes A.J.
Global metabolite profiling of human colorectal cancer xenografts in mice using HPLC-MS/MS.
J. Proteome Res. 12:2980-2986(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342; PMCID=PMC4893961
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23932154; DOI=10.1016/j.radonc.2013.06.032
Salendo J., Spitzner M., Kramer F., Zhang X., Jo P., Wolff H.A., Kitz J., Kaulfuss S., Beissbarth T., belstein M., Ghadimi M., Grade M., Gaedcke J.
Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g.
Radiother. Oncol. 108:451-457(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24042735; DOI=10.1038/oncsis.2013.35; PMCID=PMC3816225
Ahmed D., Eide P.W., Eilertsen I.A., Danielsen S.A., Eknaes M., Hektoen M., Lind G.E., Lothe R.A.
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis 2:e71.1-e71.8(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20; PMCID=PMC4178206
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=24755471; DOI=10.1158/0008-5472.CAN-14-0013
Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., O'Connor L., Wilding J.L., Bicknell D.C., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M.
Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer.
Cancer Res. 74:3238-3247(2014)
PubMed=24840470; DOI=10.1016/j.jprot.2014.05.002
Chik J.H.L., Zhou J., Moh E.S.X., Christopherson R., Clarke S.J., Molloy M.P., Packer N.H.
Comprehensive glycomics comparison between colon cancer cell cultures and tumours: implications for biomarker studies.
J. Proteomics 108:146-162(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25926053; DOI=10.1038/ncomms8002
Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B., Veronese S., Siena S., Sartore-Bianchi A., Beccuti M., Mottolese M., Linnebacher M., Cordero F., Di Nicolantonio F., Bardelli A.
The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets.
Nat. Commun. 6:7002.1-7002.10(2015)
PubMed=25944804; DOI=10.1158/1078-0432.CCR-14-2457
Bazzocco S., Dopeso H., Carton-Garcia F., Macaya I., Andretta E., Chionh F., Rodrigues P., Garrido M., Alazzouzi H., Nieto R., Sanchez A., Schwartz S. Jr., Bilic J., Mariadason J.M., Arango D.
Highly expressed genes in rapidly proliferating tumor cells as new targets for colorectal cancer treatment.
Clin. Cancer Res. 21:3695-3704(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26537799; DOI=10.1074/mcp.M115.051235; PMCID=PMC4762531
Holst S., Deuss A.J.M., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A.M., Deelder A.M., Mesker W.E., Tollenaar R.A.E.M., Rombouts Y., Wuhrer M.
N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression.
Mol. Cell. Proteomics 15:124-140(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52; PMCID=PMC4932877
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27470641; DOI=10.1021/acs.jproteome.6b00391
Guo J.-H., Cui Y.-Z., Yan Z.-Q., Luo Y.-Z., Zhang W.-L., Deng S.-Y., Tang S.-Q., Zhang G., He Q.-Y., Wang T.
Phosphoproteome characterization of human colorectal cancer SW620 cell-derived exosomes and new phosphosite discovery for C-HPP.
J. Proteome Res. 15:4060-4072(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4; PMCID=PMC5087121
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=27987026; DOI=10.1007/s00216-016-0125-5; PMCID=PMC5303640
Schunter A.J., Yue X.-S., Hummon A.B.
Phosphoproteomics of colon cancer metastasis: comparative mass spectrometric analysis of the isogenic primary and metastatic cell lines SW480 and SW620.
Anal. Bioanal. Chem. 409:1749-1763(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28683746; DOI=10.1186/s12943-017-0691-y; PMCID=PMC5498998
Berg K.C.G., Eide P.W., Eilertsen I.A., Johannessen B., Bruun J., Danielsen S.A., Bjornslett M., Meza-Zepeda L.A., Eknaes M., Lind G.E., Myklebost O., Skotheim R.I., Sveen A., Lothe R.A.
Multi-omics of 34 colorectal cancer cell lines -- a resource for biomedical studies.
Mol. Cancer 16:116.1-116.16(2017)
PubMed=28854368; DOI=10.1016/j.celrep.2017.08.010; PMCID=PMC5583477
Roumeliotis T.I., Williams S.P., Goncalves E., Alsinet C., Del Castillo Velasco-Herrera M., Aben N., Ghavidel F.Z., Michaut M., Schubert M., Price S., Wright J.C., Yu L., Yang M., Dienstmann R., Guinney J.H., Beltrao P., Brazma A., Pardo M., Stegle O., Adams D.J., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Choudhary J.S.
Genomic determinants of protein abundance variation in colorectal cancer cells.
Cell Rep. 20:2201-2214(2017)
PubMed=29101300; DOI=10.15252/msb.20177701; PMCID=PMC5731344
Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R.-S., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A., Heinzlmeir S., Johnstone E., Domingo E., Kerr D.J., Jesinghaus M., Slotta-Huspenina J., Weichert W., Knapp S., Feller S.M., Kuster B.
Pharmacoproteomic characterisation of human colon and rectal cancer.
Mol. Syst. Biol. 13:951-951(2017)
PubMed=29131639; DOI=10.1021/acs.jproteome.7b00548
Torres S., Garcia-Palmero I., Marin-Vicente C., Bartolome R.A., Calvino E., Fernandez-Acenero M.J., Casal J.I.
Proteomic characterization of transcription and splicing factors associated with a metastatic phenotype in colorectal cancer.
J. Proteome Res. 17:252-264(2018)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051; PMCID=PMC6343826
Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D.A., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F.
Differential effector engagement by oncogenic KRAS.
Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=30971826; DOI=10.1038/s41586-019-1103-9
Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., Ansari R., Harper S., Jackson D.A., McRae R., Pooley R., Wilkinson P., van der Meer D.J., Dow D., Buser-Doepner C.A., Bertotti A., Trusolino L., Stronach E.A., Saez-Rodriguez J., Yusa K., Garnett M.J.
Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens.
Nature 568:511-516(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=31978347; DOI=10.1016/j.cell.2019.12.023; PMCID=PMC7339254
Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jane-Valbuena J., Gelfand E.T., Schweppe D.K., Jedrychowski M.P., Golji J., Porter D.A., Rejtar T., Wang Y.K., Kryukov G.V., Stegmeier F., Erickson B.K., Garraway L.A., Sellers W.R., Gygi S.P.
Quantitative proteomics of the Cancer Cell Line Encyclopedia.
Cell 180:387-402.e16(2020)"