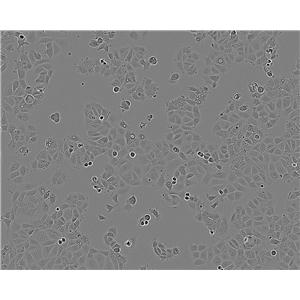
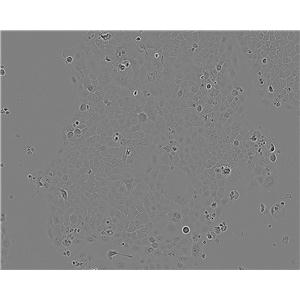
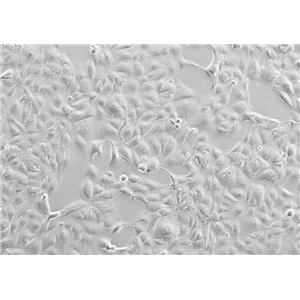
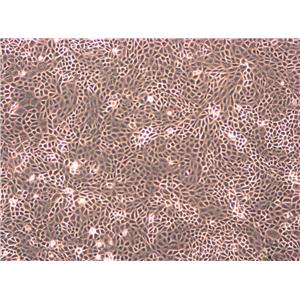
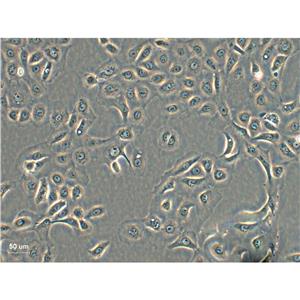
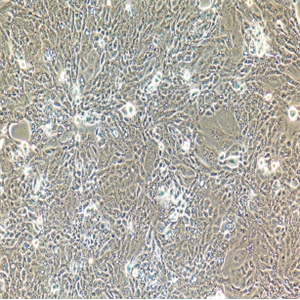
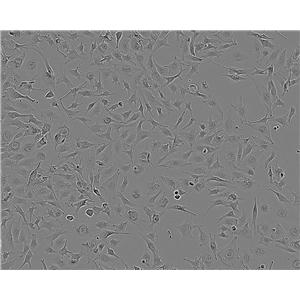
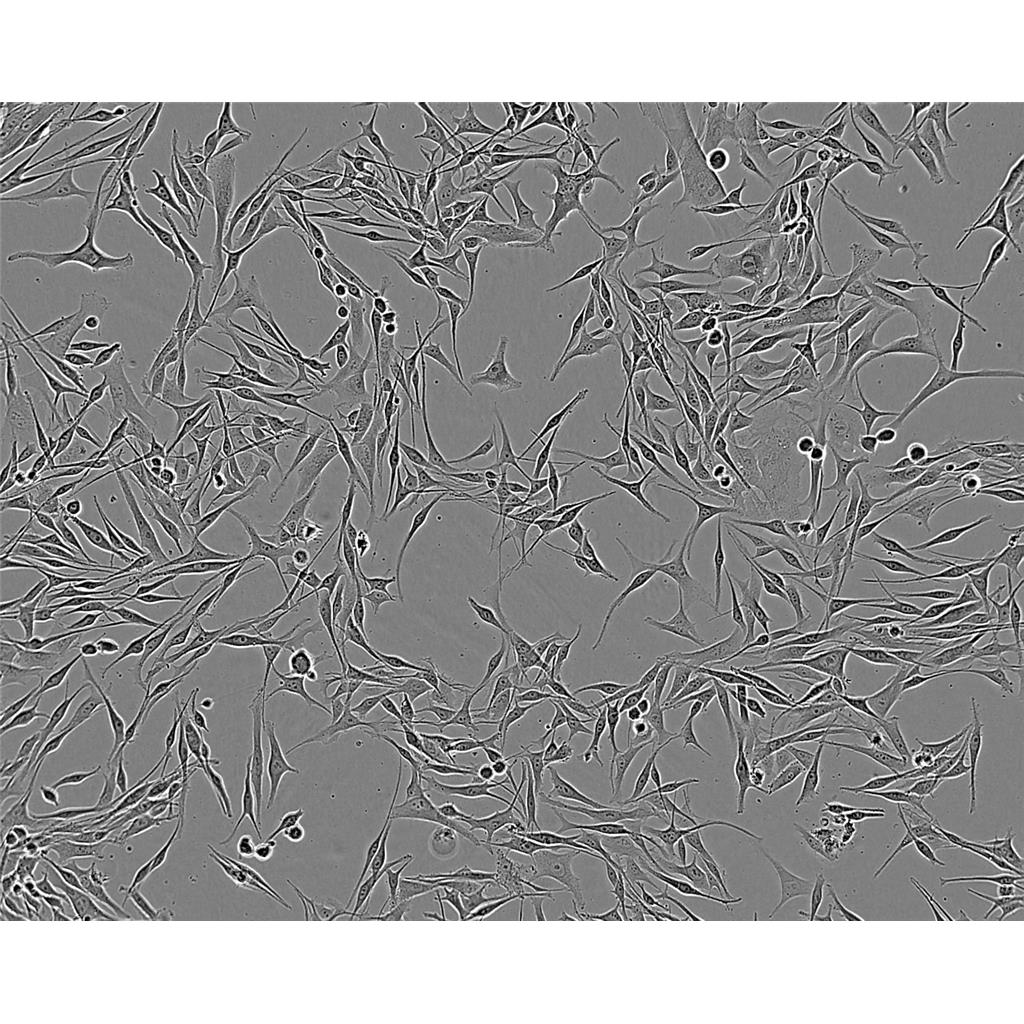
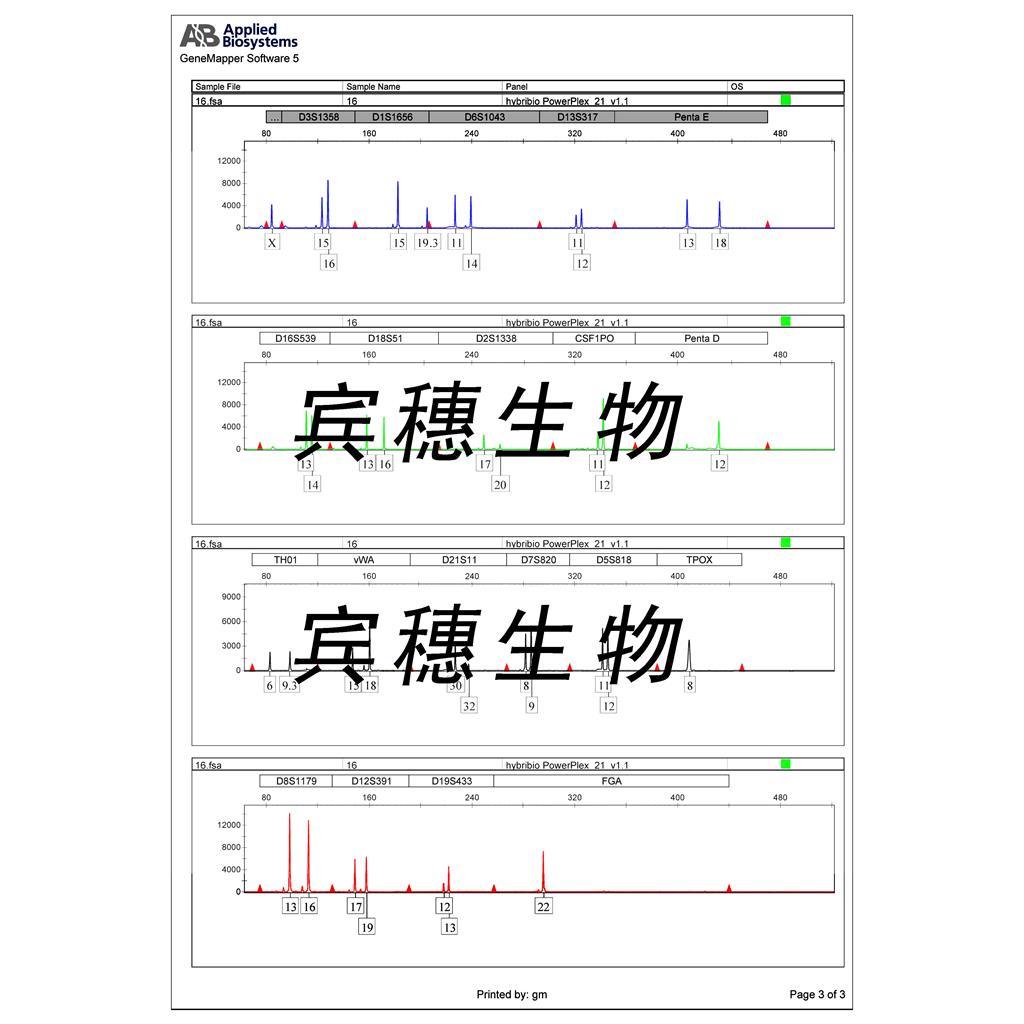
"SK-OV-3人卵巢癌细胞代次低|培养基|送STR图谱
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:贴壁生长
细胞系的选择需要考虑到细胞系的功能特点、生长速率、铺板效率、生长条件和生长特征、克隆效率、培养方式等因素,如果您想高产量表达重组蛋白,您可以选择可以悬浮生长的快速生长细胞系。细胞培养的操作步骤主要包括传代、换液、冻存和复苏。这些步骤确保了细胞能够在实验室环境中长期存活并继续增殖。传代是将细胞从一个容器转移到另一个容器的过程,以扩大细胞数量;换液是为了清除代谢废物并补充新鲜培养基;冻存则是为了长期保存细胞,而复苏则是重新激活冷冻保存的细胞使其恢复正常生长。
换液周期:每周2-3次
Ca9-22 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:C643细胞、MV-4-11细胞、NCIH128细胞
Vero 76 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SK-N-BE(1)细胞、HM细胞、PG-4 S+L-细胞
NCIH358 Cells;背景说明:1981年从一位开始化疗之前的患者的肿瘤组织中分离建株。超微结构研究表明细胞质中有Clara细胞的特征结构细胞表达主要的肺表面结合蛋白SP-A的蛋白和RNA。不表达SP-B和SP-C。他们在软琼脂中的克隆形成效率为0.83%。;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:NOMO-1细胞、Giant Cell Tumor细胞、COS-1细胞
SK-OV-3人卵巢癌细胞代次低|培养基|送STR图谱
背景信息:SK-OV-3由G.Trempe和L.J.Old在1973年从卵巢肿瘤病人的腹水分离得到。 此细胞对肿瘤坏死因子和几种细胞毒性药物包括白喉毒素、顺铂和阿霉素均耐受。 在裸鼠中致瘤,且形成与卵巢原位癌一致的中度分化的腺癌。
┈订┈购(技术服务)┈热┈线:1┈3┈6┈4┈1┈9┈3┈0┈7┈9┈1【微信同号】┈Q┈Q:3┈1┈8┈0┈8┈0┈7┈3┈2┈4;
公司细胞系主要引进ATCC、DSMZ、JCRB、KCLB、RIKEN、ECACC等细胞库,细胞系体外培养,它们会成长为单层细胞,附着或紧贴在培养瓶上,或悬浮在体外的溶液中,细胞系复苏周期短,公司细胞系状态良好,饱满,有光泽等优点。EDTA的作用:许多人不用胰酶,只用EDTA,或者用胰酶/EDTA联合作用。这里要明白,胰酶切割细胞外基质的一些负责粘连和附着的蛋白,而EDTA通过螯合Ca离子,作用于整联蛋白的活性,所以EDTA的作用更加温和。有的人在胰酶里添加一些EDTA,或者对付特别难消化的细胞,添加多一些EDTA,就是这个道理。一般不要试图延长消化时间(如果10min还消化不下来的话),而应该想其它办法。
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
SK-OV-3人卵巢癌细胞代次低|培养基|送STR图谱
物种来源:人源、鼠源等其它物种来源
HuP-T4 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:LAN-6细胞、OCI/AML-2细胞、EL4细胞
H1650_CO Cells;背景说明:该细胞是从一名27岁白人男性(10年烟龄)支气管肺泡癌患者的胸腔积液中分离得到的。;传代方法:1:4-1:6传代;2-3天换液1次。;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:SNGM细胞、SW948细胞、H295R细胞
FOX-NY Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:A-875细胞、C8161细胞、MLO-Y4细胞
L 1210 Cells;背景说明:该细胞源于用0.2%甲基胆蒽(溶解)涂抹雌性小鼠的皮肤诱发的肿瘤,鼠痘病毒阴性。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:BHP10-3细胞、NCI-SNU-5细胞、NCI-H2073细胞
┈订┈购(技术服务)┈热┈线:1┈3┈6┈4┈1┈9┈3┈0┈7┈9┈1【微信同号】┈Q┈Q:3┈1┈8┈0┈8┈0┈7┈3┈2┈4;
形态特性:上皮细胞样
贴壁消化难题:1,先用PBS 把细胞洗两遍,使瓶内没有血清了,减少对胰酶的中和,然后用新配的0.25%的胰酶加入3ml左右,放在37度,然后可以在细胞有些消化下来时,拿着瓶口,运用手腕的力量轻轻震荡瓶内体,这样细胞很快就下来了,还不需要吹打,分散也均匀;2,成团、絮状:消化里加入eda可以减少细胞成团的现象,血清可以终止胰酶的作用,如果是进口血清的话也能终止eda的作用。用胰酶消化后胰酶可以倒掉,也可以不倒,直接加血清终止,如果消化中加入了eda的话,就要将消化倒干净,如果细胞贴壁要求不是很严格的话,一般不需要进行离心。鼻咽癌细胞的贴壁能力很强,用0.5%胰酶(含0.1%EDA)一般要消化12~15min。用PBS洗涤时要洗净残余的培养基,加入胰酶后在培养箱中消化(避免细胞室温下受损以及在此温度时胰酶活性Zui强)至细胞收缩变圆(可显微镜下观察)且有少许细胞脱落(有流下来的趋势),随后立即弃去胰酶(如果脱落的细胞很多且需要大量细胞实验,则不能弃去胰酶),加入培养基仔细吹打(不能用无血清培养基或者PBS替代,否则细胞聚集成团块或絮状)。一般我都离心一次弃去上清(去除残留的胰酶及漂浮的死细胞或细胞碎片);消化过度:马上用培养基中和,用吸管吹打细胞,收集全部的细胞到以无菌的离心管中800RPM 3分钟。弃上清,用全培重悬,换新的培养瓶继续培养,状态不HAO的细胞在培养的过程中会死亡脱落,在换的时候可以清除掉!
HOS/MNNG Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-2347细胞、Centre Antoine Lacassagne-51细胞、JROECL19细胞
CAL 51 Cells;背景说明:乳腺癌;胸腔积液转移;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Bac 1.2F5细胞、Hs27细胞、L540细胞
JKT-1 Cells;背景说明:精原瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HEYA8细胞、NCI-SNU-449细胞、HCC0366细胞
P31/Fujioka Cells;背景说明:详见相关文献介绍;传代方法:1:5传代;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:B958细胞、SK BR 03细胞、D562细胞
OV1063 Cells;背景说明:卵巢上皮细胞癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:KYSE-30细胞、Colon38细胞、KMS-18细胞
Human Pancreatic Duct Epithelial Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长 ;形态特性:详见产品说明书;相关产品有:293H细胞、MDA415细胞、HPAC细胞
H1688 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:T/G HA VSMC细胞、KLM1细胞、HEL-299细胞
CCC-HEL-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SF763细胞、MDA.MB.231细胞、WM-266-mel细胞
Nb-2 Cells;背景说明:恶性淋巴瘤;雄性;Nb;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:C666-1细胞、SK MEL 5细胞、CL40细胞
RWPE2 Cells;背景说明:详见相关文献介绍;传代方法:1:3传代,2-3天传一代。;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:Stanford University-Diffuse Histiocytic Lymphoma-8细胞、DMS 153细胞、SCLC21H细胞
HNE3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:GM04678细胞、KYSE-410细胞、LM-TK-细胞
RAW264.7 Cells;背景说明:单核巨噬细胞白血病;雄性;BALB/c;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SK MEL 5细胞、U-266细胞、Panc02-H0细胞
Mouse Bladder Tumor line-2 Cells;背景说明:膀胱移行细胞癌;C3H/He;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NPA87-1细胞、Porcine Kidney-15细胞、IPLB-SF-21细胞
MAC1 Cells;背景说明:皮肤T淋巴瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:MuM-2B细胞、HCC-1428细胞、SACC-LM细胞
REC 1 Cells;背景说明:详见相关文献介绍;传代方法:1:3—1:6传代,2-3天换液1次;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:HCC-4006细胞、U266B1细胞、HOS (TE85, Clone F5)细胞
P3-X63Ag8 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HCC 2185细胞、Reuber H35细胞、H460细胞
SW 1990 Cells;背景说明:胰腺癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MLE12细胞、NB19细胞、R2C细胞
Abcam A-549 LIMK1 KO Cells(提供STR鉴定图谱)
Abcam U-87MG SDHB KO Cells(提供STR鉴定图谱)
BayGenomics ES cell line CSG163 Cells(提供STR鉴定图谱)
BayGenomics ES cell line RRT603 Cells(提供STR鉴定图谱)
BayGenomics ES cell line YTC538 Cells(提供STR鉴定图谱)
CHO Pro-3 MtxRI-6-3 Cells(提供STR鉴定图谱)
DA02389 Cells(提供STR鉴定图谱)
DD1322 Cells(提供STR鉴定图谱)
GM01893 Cells(提供STR鉴定图谱)
OACP4C Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:B-cell Acute Lymphoblastic Leukemia-1细胞、EOC 20细胞、NB-4细胞
SK-OV-3人卵巢癌细胞代次低|培养基|送STR图谱
RAMOS2G64C10 Cells;背景说明:详见相关文献介绍;传代方法: 维持细胞浓度在2×105/ml-1×106/ml;根据细胞浓度每2-3天补液1次。;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:McCoy细胞、MADB106细胞、U-87MG细胞
H-1882 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:IPECJ2细胞、K7M2细胞、MIA PaCa-2细胞
SKG-3a Cells;背景说明:详见相关文献介绍;传代方法:2x10^4 cells/ml;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:TALL1细胞、NIT-1细胞、Hs 604.T细胞
NCIH1963 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:悬浮生长;形态特性:详见产品说明书;相关产品有:C1498细胞、NR-8383细胞、ES2细胞
HEK-293-H Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:LK-2细胞、Reh细胞、SHG 44细胞
3H8-G8 Cells(提供STR鉴定图谱)
Mac-1 Cells;背景说明:皮肤T淋巴瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:U87细胞、SNU761细胞、3396细胞
NCIH1581 Cells;背景说明:详见相关文献介绍;传代方法:每周换液2次。;生长特性:混合型;形态特性:上皮样;相关产品有:H-125细胞、EB-2细胞、Soleus clone 8细胞
HCC1500 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每3-5天换液;生长特性:贴壁生长;形态特性:上皮样;相关产品有:OEC33细胞、HTR-8/SV neo细胞、H1417细胞
AC16 [Human hybrid] Cells;背景说明:心肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SU86.86细胞、L2细胞、L23/P细胞
C33 Cells;背景说明:C-33A细胞株是N. Auersperg从宫颈癌切片中建立的一系列细胞株(参见ATCC CRL-1594和ATCC CRL-1595)中的一株。 细胞一开始就表现出亚二倍体核型及上皮细胞形态。 连续传代可以观察到核型不稳定。 存在成视网膜细胞瘤蛋白(pRB),但大小不正常。 P53表达上调,且有一个273位密码子的点突变导致Arg -> Cys的替换。 人乳头瘤病毒DNA及RNA阴性。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:2B4细胞、B16-F10--RED细胞、MNNG-HOS (TE 85, clone F-5)细胞
LM8 Cells;背景说明:Dunn's骨肉瘤;雌性;C3H;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:ECV 304细胞、PK-13细胞、Centre Antoine Lacassagne-62细胞
Hs 888.T Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;每周换液2-3次;生长特性:贴壁生长;形态特性:成纤维细胞和上皮细胞的混合样;相关产品有:SPDC-CCL141细胞、LC-MS细胞、TCC-PAN2细胞
NCC-IT Cells;背景说明:详见相关文献介绍;传代方法:1:4—1:8传代,每周换液2—3次;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:NCI-H2198细胞、MGHU1细胞、SUP-T1细胞
GM28475 Cells(提供STR鉴定图谱)
HAP1 PDIK1L (-) 3 Cells(提供STR鉴定图谱)
SKMel-28 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:8传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:星形的;相关产品有:Line 207细胞、Hs 888.Lu细胞、A-498细胞
GM05372 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:J 774A.1细胞、HF-91细胞、FaDu细胞
32D.3 Cells;背景说明:骨髓淋巴瘤;C3H/HeJ;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:TKB-1细胞、Hs 636.T细胞、Capan1细胞
H1404 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:He-La细胞、Hs-343-T细胞、PT-67细胞
Cloudman S91 melanoma clone M-3 Cells;背景说明:黑色素瘤;雄性;DBA;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:451Lu细胞、BNL CL.2细胞、HBL-1 [Human diffuse large B-cell lymphoma]细胞
Tu177 Cells;背景说明:喉鳞癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:CMT64细胞、Mhh-Call 2细胞、HPDE细胞
COS1 Cells;背景说明:该细胞源自CV-1细胞株,经转染编码野生型T抗原、起始点缺陷突变的SV40得到;细胞中整合有SV40基因组完整早期区段的单个拷贝。该细胞表达T抗原,适用于需要SV40T抗原表达的载体的转染;保留SV40溶细胞性生长的特性;支持40℃时温度敏感性A209病毒的复制;支持早期区段缺失的SV40纯体的复制。因含有SV40的DNA序列,该细胞需要在2级生物安全柜中操作。;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:Pa14C细胞、H-1373细胞、YT细胞
HCM Cells;背景说明:心肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:HEK-AD293细胞、AG 9细胞、WIL2-S细胞
HS839 Cells(提供STR鉴定图谱)
KOLF2.1J PRKN R42P SNV/SNV Cells(提供STR鉴定图谱)
MOCL7 Cells(提供STR鉴定图谱)
NWR 5765 iPSC-c506 Cells(提供STR鉴定图谱)
RG-285 Cells(提供STR鉴定图谱)
UABCATe007-A Cells(提供STR鉴定图谱)
UMGi158-A-1.1 Cells(提供STR鉴定图谱)
HAP1 SLIT2 (-) 1 Cells(提供STR鉴定图谱)
OCI Ly3 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:Tokyo Medical and Dental university 8细胞、MDAMB436细胞、CESS细胞
FHL 124 Cells;背景说明:晶状体上皮;自发永生;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:38C13细胞、L-M[TK-]细胞、FRO 81-2细胞
Rat Basophilic Leukemia-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Sp2/O-Ag14细胞、PG-LH7细胞、CAL-27细胞
NPA 87 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:L-6TG细胞、32Dcl3细胞、Swiss 3T3细胞
P3X63Ag8-6-5-3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SUM102PT细胞、RINm5F细胞、RT112细胞
P3X63Ag8-6-5-3 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SUM102PT细胞、RINm5F细胞、RT112细胞
NS-1-Ag4-1 Cells;背景说明:这是P3X63Ag8(ATCCTIB-9)的一个不分泌克隆。Kappa链合成了但不分泌。能抗0.1mM8-氮杂鸟嘌呤但不能在HAT培养基中生长。据报道它是由于缺失了3-酮类固醇还原酶活性的胆固醇营养缺陷型。检测表明肢骨发育畸形病毒(鼠痘)阴性。;传代方法:1:2传代,3天内可长满。;生长特性:悬浮生长;形态特性:淋巴母细胞;相关产品有:H661细胞、Tb1Lu细胞、OUMS27细胞
Mel624 Cells;背景说明:黑色素瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NCIH847细胞、CAL 148细胞、SK Mel 24细胞
HIT Cells;背景说明:胰岛β细胞;SV40转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SUM-52细胞、PLA801C细胞、MEL-526细胞
Huh7.5 Cells;背景说明:肝癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Hs819.T细胞、L-WRN细胞、Mono Mac6细胞
P815 Cells;背景说明:肥大细胞瘤;雄性;DBA/2;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:SKMEL2细胞、NPA'87细胞、hFOB 1.19细胞
Pa18C Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:HS-766T细胞、BSC40细胞、NCI-441细胞
U266BL Cells;背景说明:详见相关文献介绍;传代方法:1:3传代,2-3天传一代;生长特性:悬浮生长 ;形态特性:淋巴母细胞样;相关产品有:HP615细胞、RMG1细胞、NOZ细胞
Normal Rat, August 3, 1983 Cells;背景说明:NR8383(正常大鼠,1983年8月3日)来源于肺灌洗时的正常大鼠肺泡巨噬细胞。细胞在gerbil肺细胞连续培养液存在下培养了大约8-9个月。随后,不再需要外源生长因子。通过有限稀释法从单个细胞克隆并亚克隆NR8383细胞,并三次用软琼脂亚克隆。细胞表现出巨噬细胞的特性,吞噬酵母多糖和铜绿,非特异性脂酶活性,Fc受体,氧化降解;分泌IL-1,TNFbeta和IL-6,可重复地响应外源生长因子。NR8383细胞响应博莱霉素,分泌TGFbeta前体。在博莱霉素刺激下,TGFbe;传代方法:1:2传代;生长特性:半贴壁生长;形态特性:巨噬细胞;相关产品有:HL-60 clone15细胞、Rat 1细胞、HOK细胞
GEO Cells;背景说明:结肠癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NCI-H1373细胞、BSC-40细胞、OCI-LY-7细胞
STPLF-fE Cells(提供STR鉴定图谱)
Kobe university Oral Squamous Cell culture-2 Cells;背景说明:详见相关文献介绍;传代方法:2.2 x 10^4 cells/ml ;生长特性:贴壁生长;形态特性:上皮样;相关产品有:SHEE细胞、NCI-H660细胞、NS-1-Ag4-1细胞
H-2052 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:6传代;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:SUM149-PT细胞、SCL II细胞、Hs 863.T细胞
SV40-MES13 Cells;背景说明:肾小球系膜;SV40转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:KRC Y细胞、G519细胞、149 PT细胞
SK-MEL3 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:5传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:D283MED细胞、MDA-MB231细胞、PaTu 8988s细胞
OCI-Ly7 Cells;背景说明:弥漫大B淋巴瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NRK 49F细胞、HFT 8810细胞、PC-2 [Human pancreatic carcinoma]细胞
BC-020 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:C-8161细胞、Vero E6细胞、B-16细胞
SK-OV-3人卵巢癌细胞代次低|培养基|送STR图谱
2780CP Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:KOPN8细胞、TFH细胞、CHP-100细胞
MASMC Cells;背景说明:气道平滑肌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MDA-MB-468-RED细胞、K562/ADR细胞、RPMI8226细胞
COLO320 DM Cells;背景说明:该细胞可产生5-羟色胺、去甲、、ACTH和甲状旁腺素。角蛋白、波形蛋白弱阳性。培养条件: RPMI 1640 10%FBS;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮+贴壁;形态特性:淋巴细胞;相关产品有:RCC 7860细胞、A431/P细胞、MDA-1386细胞
SKG-3a Cells;背景说明:详见相关文献介绍;传代方法:2x10^4 cells/ml;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:TALL1细胞、SKCO-1细胞、Hs746T细胞
SKNBE Cells;背景说明:1972年11月从一们多次化疗及放疗的扩散性神经母细胞瘤患儿骨髓穿刺物中建立了SK-N-BE(2)神经母细胞瘤细胞株。 该细胞显示中等水平的多巴胺-β-羟基酶活性。 有报道称SK-N-BE(2)细胞的饱和浓度超过1x106细胞/平方厘米。细胞形态多样,有的有长突触,有的呈上皮细胞样。 细胞会聚集,形成团块并浮起;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:PZ-HPV-7细胞、NCIH2073细胞、MGH-UI细胞
DHL-4 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:淋巴母细胞;相关产品有:NTHY-ORI3.1细胞、Mink Lung细胞、MOLM13细胞
Human Embryonic Skin Cells;背景说明:皮肤;成纤维 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:PC10细胞、Fetal Bovine Heart Endothelial细胞、NCIH345细胞
NRK-52E Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:NOR 10细胞、Vero E-6细胞、COLO320DM细胞
BayGenomics ES cell line KST302 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XC104 Cells(提供STR鉴定图谱)
CPTC-APEX1-2 Cells(提供STR鉴定图谱)
MANNUT12-4B12 Cells(提供STR鉴定图谱)
S5E5 Cells(提供STR鉴定图谱)
HOKUG Cells(提供STR鉴定图谱)
" "PubMed=6582512; DOI=10.1073/pnas.81.2.568; PMCID=PMC344720
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=4016745
Buick R.N., Pullano R., Trent J.M.
Comparative properties of five human ovarian adenocarcinoma cell lines.
Cancer Res. 45:3668-3676(1985)
PubMed=3518877; DOI=10.3109/07357908609038260
Fogh J.
Human tumor lines for cancer research.
Cancer Invest. 4:157-184(1986)
PubMed=3804493; DOI=10.1002/ijc.2910390216
Hill B.T., Whelan R.D.H., Gibby E.M., Sheer D., Hosking L.K., Shellard S.A., Rupniak H.T.
Establishment and characterisation of three new human ovarian carcinoma cell lines and initial evaluation of their potential in experimental chemotherapy studies.
Int. J. Cancer 39:219-225(1987)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=2653399; DOI=10.1038/bjc.1989.108; PMCID=PMC2247140
Hills C.A., Kelland L.R., Abel G., Siracky J., Wilson A.P., Harrap K.R.
Biological properties of ten human ovarian carcinoma cell lines: calibration in vitro against four platinum complexes.
Br. J. Cancer 59:527-534(1989)
PubMed=1892748; DOI=10.1038/bjc.1991.279; PMCID=PMC1977519
Mistry P., Kelland L.R., Abel G., Sidhar S., Harrap K.R.
The relationships between glutathione, glutathione-S-transferase and cytotoxicity of platinum drugs and melphalan in eight human ovarian carcinoma cell lines.
Br. J. Cancer 64:215-220(1991)
PubMed=2041050; DOI=10.1093/jnci/83.11.757
Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K.D., Vistica D.T., Hose C.D., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R.
Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines.
J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=1423261
Mistry P., Kelland L.R., Loh S.Y., Abel G., Murrer B.A., Harrap K.R.
Comparison of cellular accumulation and cytotoxicity of cisplatin with that of tetraplatin and amminedibutyratodichloro(cyclohexylamine)platinum(IV) (JM221) in human ovarian carcinoma cell lines.
Cancer Res. 52:6188-6193(1992)
PubMed=7718330; DOI=10.1016/0959-8049(94)00472-H
Baguley B.C., Marshall E.S., Whittaker J.R., Dotchin M.C., Nixon J., McCrystal M.R., Finlay G.J., Matthews J.H.L., Holdaway K.M., van Zijl P.L.
Resistance mechanisms determining the in vitro sensitivity to paclitaxel of tumour cells cultured from patients with ovarian cancer.
Eur. J. Cancer 31A:230-237(1995)
PubMed=8557231; DOI=10.1006/gyno.1996.0014
Skilling J.S., Squatrito R.C., Connor J.P., Niemann T., Buller R.E.
p53 gene mutation analysis and antisense-mediated growth inhibition of human ovarian carcinoma cell lines.
Gynecol. Oncol. 60:72-80(1996)
PubMed=9041185
Johnson S.W., Laub P.B., Beesley J.S., Ozols R.F., Hamilton T.C.
Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines.
Cancer Res. 57:850-856(1997)
PubMed=9554442; DOI=10.1093/jnci/90.8.597
Sabichi A.L., Hendricks D.T., Bober M.A., Birrer M.J.
Retinoic acid receptor beta expression and growth inhibition of gynecologic cancer cells by the synthetic retinoid N-(4-hydroxyphenyl) retinamide.
J. Natl. Cancer Inst. 90:597-605(1998)
PubMed=9698466; DOI=10.1006/gyno.1998.5039
Maxwell G.L., Risinger J.I., Tong B., Shaw H., Barrett J.C., Berchuck A., Futreal P.A.
Mutation of the PTEN tumor suppressor gene is not a feature of ovarian cancers.
Gynecol. Oncol. 70:13-16(1998)
PubMed=10318951; DOI=10.1073/pnas.96.10.5722; PMCID=PMC21927
Lau K.-M., Mok S.C., Ho S.-M.
Expression of human estrogen receptor-alpha and -beta, progesterone receptor, and androgen receptor mRNA in normal and malignant ovarian epithelial cells.
Proc. Natl. Acad. Sci. U.S.A. 96:5722-5727(1999)
PubMed=10700174; DOI=10.1038/73432
Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V.R., Jeffrey S.S., van de Rijn M., Waltham M.C., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O.
Systematic variation in gene expression patterns in human cancer cell lines.
Nat. Genet. 24:227-235(2000)
PubMed=10972993; DOI=10.1002/1098-2744(200008)28:4<236::AID-MC6>3.0.CO;2-H
Rauh-Adelmann C., Lau K.-M., Sabeti N., Long J.P., Mok S.C., Ho S.-M.
Altered expression of BRCA1, BRCA2, and a newly identified BRCA2 exon 12 deletion variant in malignant human ovarian, prostate, and breast cancer cell lines.
Mol. Carcinog. 28:236-246(2000)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=11565033; DOI=10.1038/35095068
Moberg K.H., Bell D.W., Pronk-Wahrer D.C.R., Haber D.A., Hariharan I.K.
Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines.
Nature 413:311-316(2001)
PubMed=11789735; DOI=10.1309/4NCM-QJ9W-QM0J-6QJE
Rhodes A., Jasani B., Couturier J., McKinley M.J., Morgan J.M., Dodson A.R., Navabi H., Miller K.D., Balaton A.J.
A formalin-fixed, paraffin-processed cell line standard for quality control of immunohistochemical assay of HER-2/neu expression in breast cancer.
Am. J. Clin. Pathol. 117:81-89(2002)
PubMed=11793438; DOI=10.1002/gcc.1221
Rao P.H., Harris C.P., Lu X.-Y., Li X.-N., Mok S.C., Lau C.C.
Multicolor spectral karyotyping of serous ovarian adenocarcinoma.
Genes Chromosomes Cancer 33:123-132(2002)
PubMed=12080474; DOI=10.1038/sj.onc.1205542
Manzano R.G., Montuenga L.M., Dayton M.A., Dent P., Kinoshita I., Vicent S., Gardner G.J., Nguyen P., Choi Y.-H., Trepel J.B., Auersperg N., Birrer M.J.
CL100 expression is down-regulated in advanced epithelial ovarian cancer and its re-expression decreases its malignant potential.
Oncogene 21:4435-4447(2002)
PubMed=12417041; DOI=10.1111/j.1349-7006.2002.tb01213.x; PMCID=PMC5926887
Watanabe T., Imoto I., Katahira T., Hirasawa A., Ishiwata I., Emi M., Takayama M., Sato A., Inazawa J.
Differentially regulated genes as putative targets of amplifications at 20q in ovarian cancers.
Jpn. J. Cancer Res. 93:1114-1122(2002)
PubMed=12960427; DOI=10.1091/mbc.E03-05-0279; PMCID=PMC266758
Schaner M.E., Ross D.T., Ciaravino G., Sorlie T., Troyanskaya O.G., Diehn M., Wang Y.C., Duran G.E., Sikic T.L., Caldeira S., Skomedal H., Tu I.-P., Hernandez-Boussard T., Johnson S.W., O'Dwyer P.J., Fero M.J., Kristensen G.B., Borresen-Dale A.-L., Hastie T.J., Tibshirani R., van de Rijn M., Teng N.N.H., Longacre T.A., Botstein D., Brown P.O., Sikic B.I.
Gene expression patterns in ovarian carcinomas.
Mol. Biol. Cell 14:4376-4386(2003)
PubMed=15748285; DOI=10.1186/1479-5876-3-11; PMCID=PMC555742
Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. 3rd, Stroncek D., Marincola F.M.
HLA class I and II genotype of the NCI-60 cell lines.
J. Transl. Med. 3:11.1-11.8(2005)
PubMed=16380993; DOI=10.1002/ijc.21671
Huang K.-C., Park D.C., Ng S.-K., Lee J.Y., Ni X.-Y., Ng W.-C., Bandera C.A., Welch W.R., Berkowitz R.S., Mok S.C., Ng S.-W.
Selenium binding protein 1 in ovarian cancer.
Int. J. Cancer 118:2433-2440(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=17726699; DOI=10.1002/gcc.20492
Buys T.P.H., Chari R., Lee E.H.-L., Zhang M., MacAulay C., Lam S., Lam W.L., Ling V.
Genetic changes in the evolution of multidrug resistance for cultured human ovarian cancer cells.
Genes Chromosomes Cancer 46:1069-1079(2007)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921; PMCID=PMC4020356
Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N.
DNA fingerprinting of the NCI-60 cell line panel.
Mol. Cancer Ther. 8:713-724(2009)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20204287; DOI=10.3892/or_00000728; PMCID=PMC2909445
Langland G.T., Yannone S.M., Langland R.A., Nakao A., Guan Y.-H., Long S.B.T., Vonguyen L., Chen D.J., Gray J.W., Chen F.-Q.
Radiosensitivity profiles from a panel of ovarian cancer cell lines exhibiting genetic alterations in p53 and disparate DNA-dependent protein kinase activities.
Oncol. Rep. 23:1021-1026(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=21912889; DOI=10.1007/s10637-011-9744-z
Sutherland H.S., Hwang I.Y., Marshall E.S., Lindsay B.S., Denny W.A., Gilchrist C., Joseph W.R., Greenhalgh D., Richardson E., Kestell P., Ding A., Baguley B.C.
Therapeutic reactivation of mutant p53 protein by quinazoline derivatives.
Invest. New Drugs 30:2035-2045(2012)
PubMed=22328975; DOI=10.1158/2159-8290.CD-11-0170; PMCID=PMC3274821
Hanrahan A.J., Schultz N., Westfal M.L., Sakr R.A., Giri D.D., Scarperi S., Janakiraman M., Olvera N., Stevens E.V., She Q.-B., Aghajanian C., King T.A., de Stanchina E., Spriggs D.R., Heguy A., Taylor B.S., Sander C., Rosen N., Levine D.A., Solit D.B.
Genomic complexity and AKT dependence in serous ovarian cancer.
Cancer Discov. 2:56-67(2012)
PubMed=22336246; DOI=10.1016/j.bmc.2012.01.017
Kong D.-X., Yamori T.
JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs.
Bioorg. Med. Chem. 20:1947-1951(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096; PMCID=PMC3285665
Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T.
Identification of cancer cell-line origins using fluorescence image-based phenomic screening.
PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22585861; DOI=10.1158/2159-8290.CD-11-0224; PMCID=PMC5057396
Marcotte R., Brown K.R., Suarez Saiz F.J., Sayad A., Karamboulas K., Krzyzanowski P.M., Sircoulomb F., Medrano M., Fedyshyn Y., Koh J.L.-Y., van Dyk D., Fedyshyn B., Luhova M., Brito G.C., Vizeacoumar F.J., Vizeacoumar F.S., Datti A., Kasimer D., Buzina A., Mero P., Misquitta C., Normand J., Haider M., Ketela T., Wrana J.L., Rottapel R., Neel B.G., Moffat J.
Essential gene profiles in breast, pancreatic, and ovarian cancer cells.
Cancer Discov. 2:172-189(2012)
PubMed=22628656; DOI=10.1126/science.1218595; PMCID=PMC3526189
Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K.
Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation.
Science 336:1040-1044(2012)
PubMed=22705003; DOI=10.1016/j.humpath.2012.03.011
Rahman M., Nakayama K., Rahman M.T., Nakayama N., Ishikawa M., Katagiri A., Iida K., Nakayama S., Otsuki Y., Shih I.-M., Miyazaki K.
Clinicopathologic and biological analysis of PIK3CA mutation in ovarian clear cell carcinoma.
Hum. Pathol. 43:2197-2206(2012)
PubMed=22710073; DOI=10.1016/j.ygyno.2012.06.017; PMCID=PMC3432677
Korch C.T., Spillman M.A., Jackson T.A., Jacobsen B.M., Murphy S.K., Lessey B.A., Jordan V.C., Bradford A.P.
DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination.
Gynecol. Oncol. 127:241-248(2012)
PubMed=23415752; DOI=10.1016/j.molonc.2012.12.007; PMCID=PMC4106023
Stordal B., Timms K., Farrelly A., Gallagher D., Busschots S., Renaud M., Thery J., Williams D., Potter J., Tran T., Korpanty G., Cremona M., Carey M.S., Li J., Li Y., Aslan O., O'Leary J.J., Mills G.B., Hennessy B.T.
BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation.
Mol. Oncol. 7:567-579(2013)
PubMed=23839242; DOI=10.1038/ncomms3126; PMCID=PMC3715866
Domcke S., Sinha R., Levine D.A., Sander C., Schultz N.
Evaluating cell lines as tumour models by comparison of genomic profiles.
Nat. Commun. 4:2126.1-2126.10(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342; PMCID=PMC4893961
Abaan O.D., Polley E.C., Davis S.R., Zhu Y.-L.J., Bilke S., Walker R.L., Pineda M.A., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S.
The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology.
Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018
Moghaddas Gholami A., Hahne H., Wu Z.-X., Auer F.J., Meng C., Wilhelm M., Kuster B.
Global proteome analysis of the NCI-60 cell line panel.
Cell Rep. 4:609-620(2013)
PubMed=24023729; DOI=10.1371/journal.pone.0072162; PMCID=PMC3762837
Anglesio M.S., Wiegand K.C., Melnyk N., Chow C., Salamanca C.M., Prentice L.M., Senz J., Yang W., Spillman M.A., Cochrane D.R., Shumansky K., Shah S.P., Kalloger S.E., Huntsman D.G.
Type-specific cell line models for type-specific ovarian cancer research.
PLoS ONE 8:E72162-E72162(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20; PMCID=PMC4178206
Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A.
The metabolic demands of cancer cells are coupled to their size and protein synthesis rates.
Cancer Metab. 1:20.1-20.13(2013)
PubMed=24434149; DOI=10.1016/j.bbrc.2013.12.070
Sinha A., Ignatchenko V., Ignatchenko A., Mejia-Guerrero S., Kislinger T.
In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome.
Biochem. Biophys. Res. Commun. 445:694-701(2014)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=25230021; DOI=10.1371/journal.pone.0103988; PMCID=PMC4167545
Beaufort C.M., Helmijr J.C.A., Piskorz A.M., Hoogstraat M., Ruigrok-Ritstier K., Besselink N., Murtaza M., van IJcken W.F.J., Heine A.A.J., Smid M., Koudijs M.J., Brenton J.D., Berns E.M.J.J., Helleman J.
Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes.
PLoS ONE 9:E103988-E103988(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25846456; DOI=10.3892/ijo.2015.2951
Takenaka M., Saito M., Iwakawa R., Yanaihara N., Saito M., Kato M., Ichikawa H., Shibata T., Yokota J., Okamoto A., Kohno T.
Profiling of actionable gene alterations in ovarian cancer by targeted deep sequencing.
Int. J. Oncol. 46:2389-2398(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=26169745; DOI=10.1186/s12967-015-0576-z; PMCID=PMC4499939
Halama A., Guerrouahen B.S., Pasquier J., Diboun I., Karoly E.D., Suhre K., Rafii A.
Metabolic signatures differentiate ovarian from colon cancer cell lines.
J. Transl. Med. 13:223.1-223.12(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=26668597; DOI=10.3892/etm.2015.2836; PMCID=PMC4665172
Ruan Z.-Y., Liu J.-H., Kuang Y.-P.
Isolation and characterization of side population cells from the human ovarian cancer cell line SK-OV-3.
Exp. Ther. Med. 10:2071-2078(2015)
PubMed=26925792; DOI=10.4149/315_150930N510
Saczko J., Pilat J., Choromanska A., Rembialkowska N., Bar J.K., Deszcz I., Zalewski J., Kulbacka J.
The effectiveness of chemotherapy and electrochemotherapy on ovarian cell lines in vitro.
Neoplasma 63:450-455(2016)
PubMed=27235858; DOI=10.1016/j.ygyno.2016.05.028; PMCID=PMC4961516
Hernandez L., Kim M.K., Lyle L.T., Bunch K.P., House C.D., Ning F., Noonan A.M., Annunziata C.M.
Characterization of ovarian cancer cell lines as in vivo models for preclinical studies.
Gynecol. Oncol. 142:332-340(2016)
PubMed=27377824; DOI=10.1038/sdata.2016.52; PMCID=PMC4932877
Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J.
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR.
Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4; PMCID=PMC5087121
Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H.
A map of mobile DNA insertions in the NCI-60 human cancer cell panel.
Mob. DNA 7:20.1-20.11(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=28273451; DOI=10.1016/j.celrep.2017.02.028
Medrano M., Communal L., Brown K.R., Iwanicki M., Normand J., Paterson J., Sircoulomb F., Krzyzanowski P.M., Novak M., Doodnauth S.A., Suarez Saiz F.J., Cullis J., Al-awar R., Neel B.G., McPherson J., Drapkin R.I., Ailles L., Mes-Masson A.-M., Rottapel R.
Interrogation of functional cell-surface markers identifies CD151 dependency in high-grade serous ovarian cancer.
Cell Rep. 18:2343-2358(2017)
PubMed=30485824; DOI=10.1016/j.celrep.2018.10.096; PMCID=PMC6481945
Papp E., Hallberg D., Konecny G.E., Bruhm D.C., Adleff V., Noe M., Kagiampakis I., Palsgrove D., Conklin D., Kinose Y., White J.R., Press M.F., Drapkin R.I., Easwaran H., Baylin S.B., Slamon D.J., Velculescu V.E., Scharpf R.B.
Integrated genomic, epigenomic, and expression analyses of ovarian cancer cell lines.
Cell Rep. 25:2617-2633(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)"