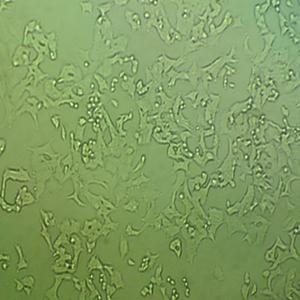
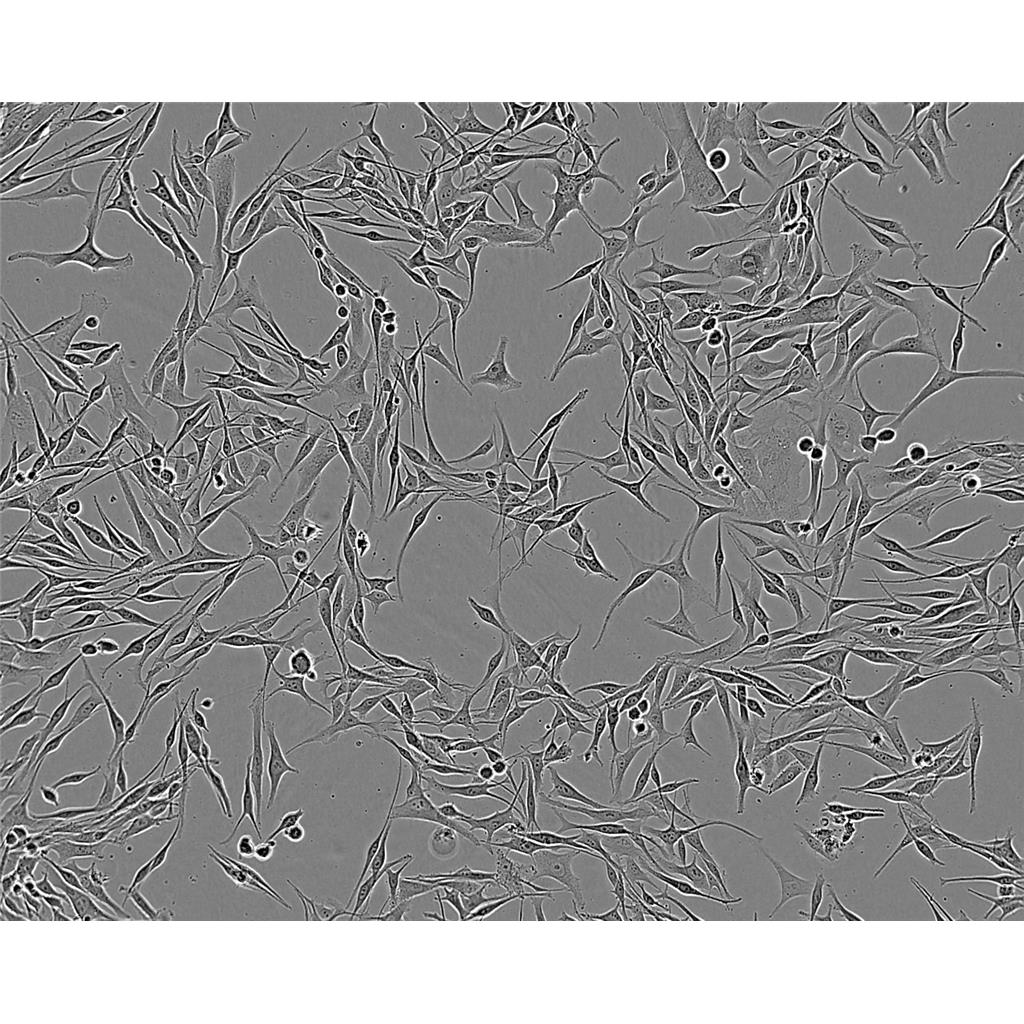
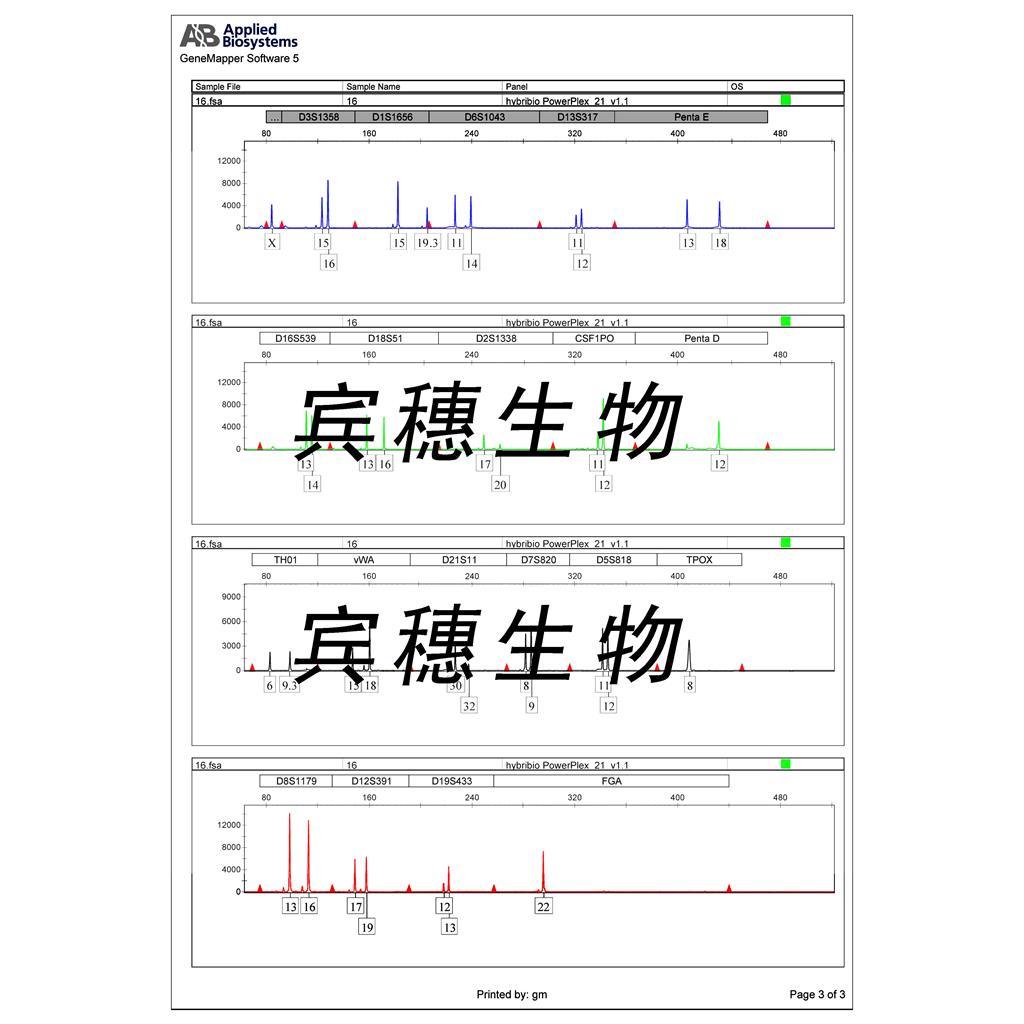
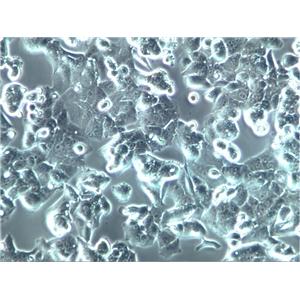
"HL-60人原髓细胞白血病细胞代次低|培养基|送STR图谱
传代比例:1:2-1:4(首次传代建议1:2)
生长特性:悬浮生长
细胞系的应用:1)免疫组化研究2)RNA干扰研究3)药物作用研究4)慢病毒转染研究等其它应用。细胞系通常用于实验研究,如增殖、迁移、侵袭等。细胞系在多个领域的研究中被广泛应用,包括基础医学、临床试验、药物筛选和分子生物学研究。这些研究不仅在中国,也在日本、美国和欧洲等多个国家和地区进行。
换液周期:每周2-3次
RT112 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:RS4;11细胞、P 815细胞、HaCaT细胞
Granta519 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:TE15细胞、Balb/c 3T3细胞、Ishikawa细胞
NCIH322 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:SW-1116细胞、H-865细胞、Henrietta Lacks cells细胞
HL-60人原髓细胞白血病细胞代次低|培养基|送STR图谱
背景信息:该细胞由CollinsSJ从一位患有急性早幼粒细胞性白血病的36岁白人女性的外周血中分离建立;可自发分化,或在盐、次黄嘌呤、佛波醇肉豆蔻(,TPA)、DMSO(1%to1.5%)、D和视黄的刺激下发生分化;刺激后可分泌TNF-α。该细胞具有吞噬活性和趋化反应,癌基因myc阳性,表达补体受体和FcR。
┈订┈购(技术服务)┈热┈线:1┈3┈6┈4┈1┈9┈3┈0┈7┈9┈1【微信同号】┈Q┈Q:3┈1┈8┈0┈8┈0┈7┈3┈2┈4;
DSMZ菌株保藏中心成立于1969年,是德国的国家菌种保藏中心。该中心一直致力于细菌、真菌、质粒、抗菌素、人体和动物细胞、植物病毒等的分类、鉴定和保藏工作。DSMZ菌种保藏中心是欧洲规模最大的生物资源中心,保藏有动物细胞500多株。Riken BRC成立于1920年,是英国的国家菌种保藏中心。该中心一直致力于细菌、真菌、植物病毒等的分类、鉴定和保藏工作。日本Riken BRC(Riken生物资源保藏中心)是全球三大典型培养物收集中心之一。Riken保藏中心提供了很多细胞系。在世界范围内,这些细胞系,都在医学、科学和兽医中具有重要意义。Riken生物资源中心支持了各种学术、健康、食品和兽医机构的研究工作,并在世界各地不同组织的微生物实验室和研究机构中使用。
产品包装:复苏发货:T25培养瓶(一瓶)或冻存发货:1ml冻存管(两支)
来源说明:细胞主要来源ATCC、ECACC、DSMZ、RIKEN等细胞库
HL-60人原髓细胞白血病细胞代次低|培养基|送STR图谱
物种来源:人源、鼠源等其它物种来源
C3H/10T1/2-clone8 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MSF细胞、SW-962细胞、NSC-34细胞
U-2932 Cells;背景说明:弥漫大B淋巴瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:Hs 839.T细胞、P3HR1细胞、NRK52E细胞
C17 Cells;背景说明:神经干细胞;C57BL/6 x CD-1;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MDA-453细胞、SK-MEL-31细胞、Tn5B1-4细胞
NCIH1688 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NCI-H23细胞、AGS细胞、786-0WT细胞
┈订┈购(技术服务)┈热┈线:1┈3┈6┈4┈1┈9┈3┈0┈7┈9┈1【微信同号】┈Q┈Q:3┈1┈8┈0┈8┈0┈7┈3┈2┈4;
形态特性:淋巴母细胞样
细胞传代培养实验:体外培养的原代细胞或细胞株要在体外持续地培养就必须传代,以便获得稳定的细胞株或得到大量的同种细胞,并维持细胞种的延续。培养的细胞形成单层汇合以后,由于密度过大生存空间不足而引起营养枯竭,将培养的细胞分散,从容器中取出,以1:2或1:3以上的比率转移到另外的容器中进行培养,即为传代培养;细胞“一代”指从细胞接种到分离再培养的一段期间,与细胞世代或倍增不同。在一代中,细胞培增3~6次。细胞传代后,一般经过三个阶段:游离期、指数增生期和停止期。常用细胞分裂指数表示细胞增殖的旺盛程度,即细胞群的分裂相数/100个细胞。一般细胞分裂指数介于0.2%~0.5%,肿瘤细胞可达3~5%;细胞接种2~3天分裂增殖旺盛,是活力ZuiHAO时期,称指数增生期(对数生长期),适宜进行各种试验。实验步骤:1.将长成的培养细胞从二氧化碳培养箱中取出,在超净工作台中倒掉瓶内的培养,加入少许消化。(以面盖住细胞为宜),静置5~10分钟。2.在倒置镜下观察被消化的细胞,如果细胞变圆,相互之间不再连接成片,这时应立即在超净台中将消化倒掉,加入3~5ml新鲜培养,吹打,制成细胞悬。3.将细胞悬吸出2ml左右,加到另一个培养瓶中并向每个瓶中分别加3ml左右培养,盖HAO瓶塞,送回二氧化碳培养箱中,继续进行培养。一般情况,传代后的细胞在2小时左右就能附着在培养瓶壁上,2~4天就可在瓶内形成单层,需要再次进行传代。
MADB106 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:JEG3细胞、SUM159PT细胞、Hs604T细胞
SU8686 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:BRL3A细胞、PANC-28细胞、LuCL4细胞
Wayne State University-Head and Neck 13 Cells;背景说明:舌鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NU-GC-4细胞、ND7/23细胞、H1792细胞
HO-8910 Cells;背景说明:HO-8910细胞株是1994年从一位51岁的中国卵巢癌患者腹水中建立的。 转移到裸鼠中形成的肿瘤集聚与患者原病灶处的形态一致。;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:H2081细胞、LA-795细胞、SUM159PT细胞
SNG-M Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:多边形;相关产品有:SKMEL5细胞、OVCA432细胞、CCC-HIE-2细胞
P3/X63-Ag8 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:MHCC 97细胞、MOVAS细胞、HCC-366细胞
H-1781 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:RD-2细胞、CHG-5细胞、HSC4细胞
H1563 Cells;背景说明:详见相关文献介绍;传代方法:1:3-1:4传代;每周换液2次。;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:GM00346细胞、T98细胞、MT2细胞
HMEC1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:LTEP-sm 1细胞、PCI-4B细胞、MDA-MB-134 VI细胞
NCI-H508 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:293-H细胞、NCI660细胞、JVM3细胞
Mc Ardle 7777 Cells;背景说明:肝癌;雌性;Buffalo;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:PLC-PRF-5细胞、MBMEC细胞、SUDHL-6细胞
SUM159PT Cells;背景说明:乳腺癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:DMS153细胞、KMH2细胞、BJA-B-1细胞
SLK Cells;背景说明:肉瘤;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:SUM-52-PE细胞、SK_N_BE2C细胞、CAMA细胞
Hep-G2 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Emory University-2细胞、FK81细胞、CCC-HSF-1细胞
KYSE-450 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:详见产品说明书;相关产品有:MHHCALL2细胞、LA-N-1细胞、SW480细胞
BS-C-1 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:U87-MG细胞、RCC-4细胞、H-1693细胞
SK-N-BE-2 Cells;背景说明:1972年11月从一们多次化疗及放疗的扩散性神经母细胞瘤患儿骨髓穿刺物中建立了SK-N-BE(2)神经母细胞瘤细胞株。 该细胞显示中等水平的多巴胺-β-羟基酶活性。 有报道称SK-N-BE(2)细胞的饱和浓度超过1x106细胞/平方厘米。细胞形态多样,有的有长突触,有的呈上皮细胞样。 细胞会聚集,形成团块并浮起;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:ID8/MOSEC细胞、BpRc1细胞、SK-CO-1细胞
Ab90 Cells(提供STR鉴定图谱)
Abcam Raji HSPA2 KO Cells(提供STR鉴定图谱)
Ban Cells(提供STR鉴定图谱)
BayGenomics ES cell line RRR573 Cells(提供STR鉴定图谱)
BayGenomics ES cell line YTA072 Cells(提供STR鉴定图谱)
CHA-hES19 Cells(提供STR鉴定图谱)
DA01689 Cells(提供STR鉴定图谱)
DD1247 Cells(提供STR鉴定图谱)
GM01882 Cells(提供STR鉴定图谱)
RT-BM 1 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:成神经细胞;相关产品有:B95.8细胞、SK MEL-28细胞、CemT4细胞
HL-60人原髓细胞白血病细胞代次低|培养基|送STR图谱
Karpas-422 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Colon 38细胞、T.T细胞、Caco-2/ATCC细胞
Hs839T Cells;背景说明:详见相关文献介绍;传代方法:1:2传代,2-3天换液1次。;生长特性:贴壁生长;形态特性:成纤维细胞;相关产品有:U-87MG ATCC细胞、PANC0203细胞、SK-Hep1细胞
HANK1 Cells;背景说明:NK/T细胞淋巴瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:MCA-38细胞、H-1781细胞、NCI-H322细胞
PLC/PRF/5 Cells;背景说明:该细胞系分泌乙肝病毒表面抗原(HBsAg)。 此细胞系原先被支原体污染,后用BM-cycline去除支原体;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Ramos.G6.C10细胞、HEY A8细胞、OS-RC-2细胞
NIH:OVCAR-8 Cells;背景说明:卵巢癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:X63-Ag8.653细胞、QBI-HEK 293A细胞、IEC-6细胞
3H5 [Mouse hybridoma against HNK-1] Cells(提供STR鉴定图谱)
NPC-TW01 Cells;背景说明:鼻咽癌;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:Pfeiffer细胞、H1770细胞、C57/B6-L细胞
C17.2 Cells;背景说明:神经干细胞;C57BL/6 x CD-1;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:U 138 MG细胞、Eca109细胞、U266S细胞
10RGB Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:OCM1A细胞、TKB-1细胞、CEM-T4细胞
L-1210 Cells;背景说明:该细胞源于用0.2%甲基胆蒽(溶解)涂抹雌性小鼠的皮肤诱发的肿瘤,鼠痘病毒阴性。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:Cattle chondrocytes细胞、3T3J2细胞、MJ细胞
J111 Cells;背景说明:单核细胞白血病;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCIH650细胞、Hs819T细胞、University of Michigan-Urothelial Carcinoma-14细胞
32D clone3 Cells;背景说明:骨髓淋巴瘤;C3H/HeJ;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:A9细胞、4T1-A细胞、PANC-02-03细胞
Homo sapiens No. 578, tumor cells Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:HS852.T细胞、NCI-H146细胞、Giant Cell Tumor细胞
AM-38 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:NRK52E细胞、EBNA-293细胞、HN-6细胞
GM11876 Cells(提供STR鉴定图谱)
HAP1 DHTKD1 (-) 3 Cells(提供STR鉴定图谱)
IOSE-Mar Cells;背景说明:卵巢;上皮细胞;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:4T1-A细胞、GM02132细胞、SEG1细胞
LIXC-002 Cells;背景说明:详见相关文献介绍;传代方法:1:3传代,3-4天传1次;生长特性:贴壁生长;形态特性:上皮细胞样;相关产品有:B4G12细胞、MRC9细胞、M059J细胞
Colon 26 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Jiyoye (P-2003)细胞、Ramos细胞、MHCC97细胞
ARPE19 Cells;背景说明:详见相关文献介绍;传代方法:1:2传代;生长特性:贴壁生长;形态特性:上皮样;相关产品有:Balb/3T3-4-Cl31细胞、U 937细胞、IM 9细胞
HCC1588 Cells;背景说明:肺鳞癌;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-211细胞、MDA-kb2细胞、H-3255细胞
EOC 20 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:SK-N-BE(1)n细胞、HRMC细胞、CORL23细胞
HRA 19 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Rat Lung Epithelial-6-T-antigen Negative细胞、NCI-H2444细胞、L02细胞
ZYM-SVEC01 Cells;背景说明:静脉血管内皮 Cells;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:EB-1细胞、HOS-143B细胞、KTCTL-140细胞
HHUi003-B Cells(提供STR鉴定图谱)
IOSE-53 Cells(提供STR鉴定图谱)
Ma-5 Cells(提供STR鉴定图谱)
ND07920 Cells(提供STR鉴定图谱)
PF8-2-112-1 Cells(提供STR鉴定图谱)
UCSD025i-13-4 Cells(提供STR鉴定图谱)
ZR-75-1 BCAR4 clone 2B Cells(提供STR鉴定图谱)
HAP1 TET1 (-) TET2 (-) 1 Cells(提供STR鉴定图谱)
HCC-44 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:NU-GC-2细胞、BLO11细胞、TH1细胞
CHL1 Cells;背景说明:详见相关文献介绍;传代方法:1:6—1:10传代;生长特性:贴壁生长;形态特性:上皮细胞;相关产品有:MBMEC细胞、NCI-H220细胞、MDA157细胞
RPMI1788 Cells;背景说明:B淋巴细胞;EBV转化;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:悬浮;形态特性:详见产品说明书;相关产品有:NCI-H1436细胞、UM-UC14细胞、NCI-H1436细胞
PZ-HPV-7 Cells;背景说明:前列腺上皮细胞;HPV18转化;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:NCI-SNU-216细胞、174xCEM.T2细胞、HEK-293FT细胞
HOS/MNNG Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-2347细胞、Centre Antoine Lacassagne-51细胞、JROECL19细胞
HOS/MNNG Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:H-2347细胞、Centre Antoine Lacassagne-51细胞、JROECL19细胞
GM00215 Cells;背景说明:肺;自发永生;雄性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:OS-RC-2细胞、NCIH524细胞、OCI AML2细胞
KYSE-410 Cells;背景说明:食管鳞癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:TE1细胞、SKMEL-1细胞、BTT739细胞
GM16136 Cells;背景说明:肺;自发永生;雄性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:RGM1细胞、MUS-M1细胞、C3H10T1-2细胞
HCC95 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:RN-c细胞、Neuro-2a细胞、SV-HUC细胞
HO-8910 PM Cells;背景说明:高转移卵巢癌 Cells;传代方法:消化3-5分钟。1:2。3天内可长满。;生长特性:贴壁生长;形态特性:上皮样;相关产品有:CL-40细胞、H-2227细胞、NCI-H345细胞
Hos TE-85 Cells;背景说明:骨肉瘤;女性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MS1细胞、H650细胞、ACC3细胞
H-2135 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:ROS17/28细胞、SKRC20细胞、Epstein-Barr-1细胞
H69C Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:4传代,每周换液2次;生长特性:悬浮生长,聚团;形态特性:聚团悬浮;相关产品有:H522细胞、MEF (CF-1)细胞、Jurkat E6-1细胞
NCI-SNU-1040 Cells;背景说明:结肠癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:DHL-2细胞、NIE 115细胞、H-1417细胞
SES-MaBr-3 Cells(提供STR鉴定图谱)
HL-60 Clone 15 Cells;背景说明:详见相关文献介绍;传代方法:维持细胞浓度在1×105-1×106/ml,每2-3天换液1次。;生长特性:悬浮生长;形态特性:淋巴母细胞样;相关产品有:HHUA细胞、Roswell Park Memorial Institute 7666细胞、TE-6细胞
SNUC2A Cells;背景说明:详见相关文献介绍;传代方法:每周两次换液;生长特性:松散附着、多单元的聚合;形态特性:上皮细胞样;相关产品有:A 549细胞、Panc_02_03细胞、Michigan Cancer Foundation-12A细胞
COLO 824 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:KY-270细胞、NP-69细胞、HEK293-EBNA细胞
SuperTube Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H1930细胞、TGW-I-nu细胞、LM6细胞
MRC 5 Cells;背景说明:MRC-5细胞系来自14周龄男性胎儿的正常肺组织,该细胞老化前能传代42~46个倍增时间。;传代方法:1:2-1:5传代;每周1-2次。;生长特性:贴壁生长;形态特性:成纤维细胞样;相关产品有:BE2-C细胞、PE CA PJ34细胞、RC-4B细胞
FOX-NY Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:A-875细胞、C8161细胞、MLO-Y4细胞
HL-60人原髓细胞白血病细胞代次低|培养基|送STR图谱
Huh 7.5 Cells;背景说明:肝癌;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:MiaPaCa2细胞、Lewis Lung Cancer细胞、SUM 190细胞
SKRC-52 Cells;背景说明:肾癌;纵隔膜转移;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:VA-ES-BJ细胞、Ly19细胞、Sci-1细胞
DoTc2 Cells;背景说明:详见相关文献介绍;传代方法:1:2—1:3传代,每周换液2—3次;生长特性:贴壁生长 ;形态特性:上皮样;相关产品有:Hs 688(A).T细胞、FU-OV-1细胞、MV4:11细胞
HRA 19 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:Rat Lung Epithelial-6-T-antigen Negative细胞、FC33细胞、SVG(P12)细胞
LN382 Cells;背景说明:胶质瘤;男性;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁;形态特性:详见产品说明书;相关产品有:526mel细胞、NCI-H28细胞、LN-382细胞
L-363 Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:3传代;每周换液2-3次。;生长特性:贴壁或悬浮,详见产品说明书部分;形态特性:详见产品说明书;相关产品有:H2342细胞、TE-7细胞、DoTc2细胞
HEK293FT Cells;背景说明:该细胞稳定表达SV40大T抗原,并且促进最适病毒产物的产生。;传代方法:1:2传代;生长特性:悬浮生长;形态特性:圆形;相关产品有:Evsa T细胞、SUM-52PE细胞、Tokyo Medical and Dental university 8细胞
Malme-3 M Cells;背景说明:详见相关文献介绍;传代方法:1:2-1:4传代,2天换液1次。;生长特性:混合生长;形态特性:成纤维细胞;相关产品有:RIN-m5F细胞、MDA-MB-468细胞、SUM149PT细胞
BayGenomics ES cell line NPX025 Cells(提供STR鉴定图谱)
BayGenomics ES cell line XC366 Cells(提供STR鉴定图谱)
CPTC-DDIT3-2 Cells(提供STR鉴定图谱)
MAsg2 Cells(提供STR鉴定图谱)
SC17.122 Cells(提供STR鉴定图谱)
HPB/VP-16 Cells(提供STR鉴定图谱)
" "PubMed=6255959; DOI=10.1016/0014-4827(81)90422-5
Olsson I., Olofsson T.
Induction of differentiation in a human promyelocytic leukemic cell line (HL-60). Production of granule proteins.
Exp. Cell Res. 131:225-230(1981)
PubMed=6945469; DOI=10.1016/0145-2126(81)90018-7
Major P.P., Griffin J.D., Minden M.D., Kufe D.W.
A blast subclone of the HL-60 human promyelocytic cell line.
Leuk. Res. 5:429-430(1981)
PubMed=6954533; DOI=10.1073/pnas.79.7.2194; PMCID=PMC346157
Westin E.H., Gallo R.C., Arya S.K., Eva A., Souza L.M., Baluda M.A., Aaronson S.A., Wong-Staal F.
Differential expression of the amv gene in human hematopoietic cells.
Proc. Natl. Acad. Sci. U.S.A. 79:2194-2198(1982)
PubMed=6091813; DOI=10.1182/blood.V64.5.1059.1059
Palumbo A., Minowada J., Erikson J., Croce C.M., Rovera G.
Lineage infidelity of a human myelogenous leukemia cell line.
Blood 64:1059-1063(1984)
PubMed=6582512; DOI=10.1073/pnas.81.2.568; PMCID=PMC344720
Mattes M.J., Cordon-Cardo C., Lewis J.L. Jr., Old L.J., Lloyd K.O.
Cell surface antigens of human ovarian and endometrial carcinoma defined by mouse monoclonal antibodies.
Proc. Natl. Acad. Sci. U.S.A. 81:568-572(1984)
PubMed=2858093; DOI=10.1073/pnas.82.3.790; PMCID=PMC397132
Wolf D., Rotter V.
Major deletions in the gene encoding the p53 tumor antigen cause lack of p53 expression in HL-60 cells.
Proc. Natl. Acad. Sci. U.S.A. 82:790-794(1985)
PubMed=2985879; DOI=10.1016/0145-2126(85)90084-0
Drexler H.G., Gaedicke G., Minowada J.
Isoenzyme studies in human leukemia-lymphoma cell lines -- 1 carboxylic esterase.
Leuk. Res. 9:209-229(1985)
PubMed=3159941; DOI=10.1016/0145-2126(85)90134-1
Drexler H.G., Gaedicke G., Minowada J.
Isoenzyme studies in human leukemia-lymphoma cell lines -- III Beta-hexosaminidase (E.C. 3.2.1.30).
Leuk. Res. 9:549-559(1985)
PubMed=3860286
Bhalla K.N., Hindenburg A.A., Taub R.N., Grant S.
Isolation and characterization of an anthracycline-resistant human leukemic cell line.
Cancer Res. 45:3657-3662(1985)
PubMed=3874327; DOI=10.1016/0145-2126(85)90133-x
Drexler H.G., Gaedicke G., Minowada J.
Isoenzyme studies in human leukemia-lymphoma cells lines -- II. Acid phosphatase.
Leuk. Res. 9:537-548(1985)
PubMed=3311197; DOI=10.1182/blood.V70.5.1233.1233
Collins S.J.
The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression.
Blood 70:1233-1244(1987)
DOI=10.1159/000415060
Morikawa S., Harada T., Katoh T.
Heterogeneity of cellular origins in human malignant lymphoma cell line derived from histo-monocytic lineage cells.
(In book chapter) Cellular, molecular, genetic approaches to immunodiagnosis and immunotherapy. 8th International conference on labeled antibodies, Tokyo, November 1985; Kano K., Mori S., Sugisaki T., Torisu M. (eds.); pp.373-380; Karger; Basel; Switzerland (1988)
PubMed=3335022
Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R.
Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay.
Cancer Res. 48:589-601(1988)
PubMed=3422031; DOI=10.1182/blood.V71.1.242.242
Dalton W.T. Jr., Ahearn M.J., McCredie K.B., Freireich E.J., Stass S.A., Trujillo J.M.
HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3.
Blood 71:242-247(1988)
PubMed=2545346
Hindenburg A.A., Gervasoni J.E. Jr., Krishna S., Stewart V.J., Rosado M., Lutzky J., Bhalla K.N., Baker M.A., Taub R.N.
Intracellular distribution and pharmacokinetics of daunorubicin in anthracycline-sensitive and -resistant HL-60 cells.
Cancer Res. 49:4607-4614(1989)
PubMed=1970118; DOI=10.1128/mcb.10.5.2154-2163.1990; PMCID=PMC360563
Collins S.J., Robertson K.A., Mueller L.
Retinoic acid-induced granulocytic differentiation of HL-60 myeloid leukemia cells is mediated directly through the retinoic acid receptor (RAR-alpha).
Mol. Cell. Biol. 10:2154-2163(1990)
PubMed=2140233; DOI=10.1111/j.1440-1827.1990.tb01549.x
Nakano A., Harada T., Morikawa S., Kato Y.
Expression of leukocyte common antigen (CD45) on various human leukemia/lymphoma cell lines.
Acta Pathol. Jpn. 40:107-115(1990)
PubMed=1368402; DOI=10.1007/BF02540028
Schumpp-Vonach B., Schlaeger E.-J.
Growth study of lactate and ammonia double-resistant clones of HL-60 cells.
Cytotechnology 8:39-44(1992)
PubMed=1574572; DOI=10.2307/3578273
Dunphy E.J., Beckett M.A., Thompson L.H., Weichselbaum R.R.
Expression of the polymorphic human DNA repair gene XRCC1 does not correlate with radiosensitivity in the cells of human head and neck tumor cell lines.
Radiat. Res. 130:166-170(1992)
PubMed=8316623; DOI=10.2307/3578190
Evans H.H., Ricanati M., Horng M.-F., Jiang Q.-Y., Mencl J., Olive P.L.
DNA double-strand break rejoining deficiency in TK6 and other human B-lymphoblast cell lines.
Radiat. Res. 134:307-315(1993)
PubMed=8343448
Kajigaya Y., Sasaki H., Ikuta K., Matsuyama S., Hirabayashi Y., Inoue T.
Serum-free culture for leukemia cells.
Hum. Cell 6:49-56(1993)
PubMed=7630190
Zhou M.-X., Gu L.-B., James C.D., He J., Yeager A.M., Smith S.D., Findley H.W. Jr.
Homozygous deletions of the CDKN2 (MTS1/p16ink4) gene in cell lines established from children with acute lymphoblastic leukemia.
Leukemia 9:1159-1161(1995)
PubMed=8562479
Banerjee D., Lenz H.-J., Schnieders B., Manno D.J., Ju J.-F., Spears C.P., Hochhauser D., Danenberg K.D., Danenberg P.V., Bertino J.R.
Transfection of wild-type but not mutant p53 induces early monocytic differentiation in HL60 cells and increases their sensitivity to stress.
Cell Growth Differ. 6:1405-1413(1995)
PubMed=8558913
Morita S., Tsuchiya S., Fujie H., Itano M., Ohashi Y., Minegishi M., Imaizumi M., Endo M., Takano N., Konno T.
Cell surface c-kit receptors in human leukemia cell lines and pediatric leukemia: selective preservation of c-kit expression on megakaryoblastic cell lines during adaptation to in vitro culture.
Leukemia 10:102-105(1996)
PubMed=9434637; DOI=10.1006/excr.1997.3813
Piepmeier E.H., Kalns J.E., McIntyre K.M., Lewis M.L.
Prolonged weightlessness affects promyelocytic multidrug resistance.
Exp. Cell Res. 237:410-418(1997)
PubMed=9510473; DOI=10.1111/j.1349-7006.1998.tb00476.x; PMCID=PMC5921588
Hosoya N., Hangaishi A., Ogawa S., Miyagawa K., Mitani K., Yazaki Y., Hirai H.
Frameshift mutations of the hMSH6 gene in human leukemia cell lines.
Jpn. J. Cancer Res. 89:33-39(1998)
PubMed=9557624; DOI=10.1038/sj.leu.2400976
Mattii L., Barale R., Petrini M.
Use of the comet test in the evaluation of multidrug resistance of human cell lines.
Leukemia 12:627-632(1998)
PubMed=9607592
Ju J.-F., Banerjee D., Lenz H.-J., Danenberg K.D., Schmittgen T.D., Spears C.P., Schonthal A.H., Manno D.J., Hochhauser D., Bertino J.R., Danenberg P.V.
Restoration of wild-type p53 activity in p53-null HL-60 cells confers multidrug sensitivity.
Clin. Cancer Res. 4:1315-1322(1998)
PubMed=9737686; DOI=10.1038/sj.leu.2401112
Zhang W.-J., Ohnishi K., Shigeno K., Fujisawa S., Naito K., Nakamura S., Takeshita K., Takeshita A., Ohno R.
The induction of apoptosis and cell cycle arrest by arsenic trioxide in lymphoid neoplasms.
Leukemia 12:1383-1391(1998)
PubMed=9738977; DOI=10.1111/j.1349-7006.1998.tb03275.x; PMCID=PMC5921886
Takizawa J., Suzuki R., Kuroda H., Utsunomiya A., Kagami Y., Joh T., Aizawa Y., Ueda R., Seto M.
Expression of the TCL1 gene at 14q32 in B-cell malignancies but not in adult T-cell leukemia.
Jpn. J. Cancer Res. 89:712-718(1998)
PubMed=10739008; DOI=10.1016/S0145-2126(99)00182-4
Inoue K., Kohno T., Takakura S., Hayashi Y., Mizoguchi H., Yokota J.
Frequent microsatellite instability and BAX mutations in T cell acute lymphoblastic leukemia cell lines.
Leuk. Res. 24:255-262(2000)
DOI=10.1016/B978-0-12-221970-2.50457-5
Drexler H.G.
The leukemia-lymphoma cell line factsbook.
(In book) ISBN 9780122219702; pp.1-733; Academic Press; London; United Kingdom (2001)
PubMed=11226526; DOI=10.1016/S0145-2126(00)00121-1
Inoue K., Kohno T., Takakura S., Hayashi Y., Mizoguchi H., Yokota J.
Corrigendum to: Frequent microsatellite instability and BAX mutations in T cell acute lymphoblastic leukemia cell lines Leukemia Research 24 (2000), 255-262.
Leuk. Res. 25:275-278(2001)
PubMed=11416159; DOI=10.1073/pnas.121616198; PMCID=PMC35459
Masters J.R.W., Thomson J.A., Daly-Burns B., Reid Y.A., Dirks W.G., Packer P., Toji L.H., Ohno T., Tanabe H., Arlett C.F., Kelland L.R., Harrison M., Virmani A.K., Ward T.H., Ayres K.L., Debenham P.G.
Short tandem repeat profiling provides an international reference standard for human cell lines.
Proc. Natl. Acad. Sci. U.S.A. 98:8012-8017(2001)
PubMed=12068308; DOI=10.1038/nature00766
Davies H.R., Bignell G.R., Cox C., Stephens P.J., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C.S., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H.F., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A.
Mutations of the BRAF gene in human cancer.
Nature 417:949-954(2002)
PubMed=12661003; DOI=10.1002/gcc.10196
Seitz S., Wassmuth P., Plaschke J., Schackert H.K., Karsten U., Santibanez-Koref M.F., Schlag P.M., Scherneck S.
Identification of microsatellite instability and mismatch repair gene mutations in breast cancer cell lines.
Genes Chromosomes Cancer 37:29-35(2003)
PubMed=16120335; DOI=10.1016/S1567-7249(03)00010-2
Levin B.C., Holland K.A., Hancock D.K., Coble M., Parsons T.J., Kienker L.J., Williams D.W., Jones M., Richie K.L.
Comparison of the complete mtDNA genome sequences of human cell lines -- HL-60 and GM10742A -- from individuals with pro-myelocytic leukemia and Leber hereditary optic neuropathy, respectively, and the inclusion of HL-60 in the NIST human mitochondrial DNA standard reference material -- SRM 2392-I.
Mitochondrion 2:387-400(2003)
PubMed=15843827; DOI=10.1038/sj.leu.2403749
Andersson A., Eden P., Lindgren D., Nilsson J., Lassen C., Heldrup J., Fontes M., Borg A., Mitelman F., Johansson B., Hoglund M., Fioretos T.
Gene expression profiling of leukemic cell lines reveals conserved molecular signatures among subtypes with specific genetic aberrations.
Leukemia 19:1042-1050(2005)
PubMed=16408098; DOI=10.1038/sj.leu.2404081
Quentmeier H., MacLeod R.A.F., Zaborski M., Drexler H.G.
JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders.
Leukemia 20:471-476(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433; PMCID=PMC2705832
Ikediobi O.N., Davies H.R., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A.P., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Miroo T., Parker A., Perry J., Raine K.M., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P.J., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R.
Mutation analysis of 24 known cancer genes in the NCI-60 cell line set.
Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=20164919; DOI=10.1038/nature08768; PMCID=PMC3145113
Bignell G.R., Greenman C.D., Davies H.R., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458; PMCID=PMC2881662
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=22068913; DOI=10.1073/pnas.1111840108; PMCID=PMC3219108
Gillet J.-P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M.
Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance.
Proc. Natl. Acad. Sci. U.S.A. 108:18708-18713(2011)
PubMed=22032829; DOI=10.1016/j.mrfmmm.2011.10.005
Luan Y., Kogi M., Rajaguru P., Ren J., Yamaguchi T., Suzuki K., Suzuki T.
Microarray analysis of responsible genes in increased growth rate in the subline of HL60 (HL60RG) cells.
Mutat. Res. 731:20-26(2012)
PubMed=22347499; DOI=10.1371/journal.pone.0031628; PMCID=PMC3276511
Ruan X.-Y., Kocher J.-P.A., Pommier Y., Liu H.-F., Reinhold W.C.
Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared to HapMap trios, and relation to fragile site location.
PLoS ONE 7:E31628-E31628(2012)
PubMed=22460905; DOI=10.1038/nature11003; PMCID=PMC3320027
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=22944676; DOI=10.1515/hsz-2012-0195
Reinke S.O., Bayer M., Berger M., Hinderlich S., Blanchard V.
The analysis of N-glycans of cell membrane proteins from human hematopoietic cell lines reveals distinctions in their pattern.
Biol. Chem. 393:731-747(2012)
PubMed=23955599; DOI=10.1038/ng.2731
Kon A., Shih L.-Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y., Yoshida K.-i., Okuno Y., Bando M., Nakato R., Ishikawa S., Sato-Otsubo A., Nagae G., Nishimoto A., Haferlach C., Nowak D., Sato Y., Alpermann T., Nagasaki M., Shimamura T., Tanaka H., Chiba K., Yamamoto R., Yamaguchi T., Otsu M., Obara N., Sakata-Yanagimoto M., Nakamaki T., Ishiyama K., Nolte F., Hofmann W.-K., Miyawaki S., Chiba S., Mori H., Nakauchi H., Koeffler H.P., Aburatani H., Haferlach T., Shirahige K., Miyano S., Ogawa S.
Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms.
Nat. Genet. 45:1232-1237(2013)
PubMed=24368162; DOI=10.1016/j.exphem.2013.12.004
Sripayap P., Nagai T., Uesawa M., Kobayashi H., Tsukahara T., Ohmine K., Muroi K., Ozawa K.
Mechanisms of resistance to azacitidine in human leukemia cell lines.
Exp. Hematol. 42:294-306.e2(2014)
PubMed=24670534; DOI=10.1371/journal.pone.0092047; PMCID=PMC3966786
Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.
High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner.
PLoS ONE 9:E92047-E92047(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35; PMCID=PMC4432652
Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G.-Z., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E.T., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H.-B., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Meyers B.H., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C.
Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies.
Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080
Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z.-S., Liu H.-B., Degenhardt J., Mayba O., Gnad F., Liu J.-F., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M.-M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z.-M.
A comprehensive transcriptional portrait of human cancer cell lines.
Nat. Biotechnol. 33:306-312(2015)
PubMed=25877200; DOI=10.1038/nature14397
Yu M., Selvaraj S.K., Liang-Chu M.M.Y., Aghajani S., Busse M., Yuan J., Lee G., Peale F.V., Klijn C., Bourgon R., Kaminker J.S., Neve R.M.
A resource for cell line authentication, annotation and quality control.
Nature 520:307-311(2015)
PubMed=25894527; DOI=10.1371/journal.pone.0121314; PMCID=PMC4404347
Bausch-Fluck D., Hofmann A., Bock T., Frei A.P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R.L., Yoon C., Schiess R., Schmidt A., Mirkowska P., Hartlova A.S., Van Eyk J.E., Bourquin J.-P., Aebersold R., Boheler K.R., Zandstra P.W., Wollscheid B.
A mass spectrometric-derived cell surface protein atlas.
PLoS ONE 10:E0121314-E0121314(2015)
PubMed=26589293; DOI=10.1186/s13073-015-0240-5; PMCID=PMC4653878
Scholtalbers J., Boegel S., Bukur T., Byl M., Goerges S., Sorn P., Loewer M., Sahin U., Castle J.C.
TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression.
Genome Med. 7:118.1-118.7(2015)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017; PMCID=PMC4967469
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Miroo T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=28109323; DOI=10.1186/s13045-017-0396-0; PMCID=PMC5251306
Masetti R., Bertuccio S.N., Astolfi A., Chiarini F., Lonetti A., Indio V., De Luca M., Bandini J., Serravalle S., Franzoni M., Pigazzi M., Martelli A.M., Basso G., Locatelli F., Pession A.
Hh/Gli antagonist in acute myeloid leukemia with CBFA2T3-GLIS2 fusion gene.
J. Hematol. Oncol. 10:26.1-26.5(2017)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005; PMCID=PMC5501076
Li J., Zhao W., Akbani R., Liu W.-B., Ju Z.-L., Ling S.-Y., Vellano C.P., Roebuck P., Yu Q.-H., Eterovic A.K., Byers L.A., Davies M.A., Deng W.-L., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y.-L., Mills G.B., Liang H.
Characterization of human cancer cell lines by reverse-phase protein arrays.
Cancer Cell 31:225-239(2017)
PubMed=29491412; DOI=10.1038/s41388-018-0150-2; PMCID=PMC5955861
Gerlach D., Tontsch-Grunt U., Baum A., Popow J., Scharn D., Hofmann M.H., Engelhardt H., Kaya O., Beck J., Schweifer N., Gerstberger T., Zuber J., Savarese F., Kraut N.
The novel BET bromodomain inhibitor BI 894999 represses super-enhancer-associated transcription and synergizes with CDK9 inhibition in AML.
Oncogene 37:2687-2701(2018)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5; PMCID=PMC6167786
Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H.H., Koeffler H.P.
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.
BMC Cancer 18:940.1-940.13(2018)
PubMed=30629668; DOI=10.1371/journal.pone.0210404; PMCID=PMC6328144
Uphoff C.C., Pommerenke C., Denkmann S.A., Drexler H.G.
Screening human cell lines for viral infections applying RNA-Seq data analysis.
PLoS ONE 14:E0210404-E0210404(2019)
PubMed=30670178; DOI=10.1016/j.jcf.2018.06.007
Jennings S., Ng H.P., Wang G.-S.
Establishment of a DeltaF508-CF promyelocytic cell line for cystic fibrosis research and drug screening.
J. Cyst. Fibros. 18:44-53(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747; PMCID=PMC6445675
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3; PMCID=PMC6697103
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)"