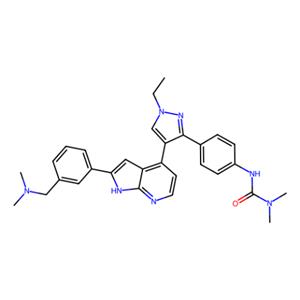
| 名称 | GSK-1070916 |
| 描述 | GSK-1070916 (GSK-1070916A) is a reversible and ATP-competitive inhibitor of Aurora B/C with IC50 of 3.5 nM/6.5 nM. It displays >100-fold selectivity against the closely related Aurora A-TPX2 complex. Phase 1. |
| 细胞实验 | Cells are plated in 96-well plates in the recommended growth media and incubated at 37 °C in 5% CO2 overnight. The following day, the cells are treated with serial dilutions of GSK1070916. At this time, one set of cells is treated with CellTiter-Glo for a time equal to 0 (T = 0) measurement. Following a 6- to 7-d incubation with compound, cell proliferation is measured using the CellTiter-Glo reagent according to the manufacture's recommended protocol. As inhibition of Aurora B induces endomitosis, the degree of which differs depending on the cell type, an extended compound treatment time is required to accurately reflect the effects on cell viability across a large panel of cell lines. For analysis of cell viability, values from wells with no cells are subtracted for background correction and the data plotted as a percent of the DMSO-treated control samples using Microsoft Excel XLfit4 software. The EC50 values represent the concentration of GSK1070916 where 50% maximal effect is observed(Only for Reference) |
| 激酶实验 | Kinase Assay: The ability of GSK1070916 to inhibit the Aurora enzymes is measured using in vivo kinase assays. The assays measure the ability of Aurora A, Aurora B and Aurora C to phosphorylate a synthetic peptide substrate. Biotin-Ahx-RARRRLSFFFFAKKK-NH2 is used for the Aurora A–TPX2 LEADseekerTM assay and 5FAM-PKAtide is used for the IMAPTM assay for all three Aurora kinases. To take into account time-dependent inhibition of Aurora enzymes, Aurora A–TPX2, Aurora B–INCENP and Aurora C–INCENP are incubated with GSK1070916 at various concentrations for 30 min before the reactions are initiated with the addition of substrates. For the Aurora A LEADseekerTM assay, final assay conditions are 0.5 nM Aurora A–TPX2, 1 μM peptide substrate, 6 mM MgCl2, 1.5 μM ATP, 0.003 μCi/μL [γ-33P] ATP in 50 mM Hepes, pH 7.2, 0.15 mg/mL BSA, 0.01% Tween-20, 5 mM DTT and 25 mM KCl. The reactions are incubated at room temperature (25 °C) for 120 min and terminated by the addition of LEADseekerTM beads in PBS containing EDTA (final concentration 2 mg/mL beads and 25 mM EDTA). The plates are then sealed, and the beads are allowed to settle overnight. Product formation is quantified using a Viewlux Imager. For the IMAPTM assays, Aurora A–TPX2 (final concentration 1 nM), Aurora B–INCENP (final concentration 2 nM) or Aurora C–INCENP (final concentration 2.5 nM) is added to the compound-containing plates in 5 μL of buffer (25 mM Hepes, pH 7.2, for Aurora A, 25 mM Hepes, pH 7.5, for Aurora B and 20 mM Hepes, pH 7.2, for Aurora C) containing 0.15 mg/mL BSA, 0.01% Tween 20 and 25 mM NaCl. This mixture is incubated at room temperature for 30 min. To start the reaction, 5 μL of a substrate solution is added containing the same Hepes buffer as used for the pre-incubation, 25 mM NaCl, MgCl2 (2, 4 and 4 mM for Aurora A, B and C respectively), DTT (4, 4 and 2 mM for Aurora A, B and C respectively), ATP (4, 4 and 10 μM for Aurora A, B and C respectively), 200 nM 5FAM-PKAtide, 0.01% Tween 20 and 0.15 mg/mL BSA. The reactions are incubated at room temperature for 120 min for Aurora A and B and 60 min for Aurora C. These reactions are then terminated by the addition of 10 μL of 1:500 (1:600 for Aurora C) Progressive Binding Reagent in 95% Progressive Binding Buffer A and 5% Progressive Binding Buffer B. Plates are incubated at room temperature for approx. 90–120 min (time allowed for equilibrium to be reached). Plates are read in a Molecular Devices Analyst plate reader in fluorescence polarization mode. |
| 体外活性 | GSK1070916 对 Aurora B和Aurora C的 Ki值为 0.38 nM和1.5 nM。针对Aurora B和C的抑制作用是时间依赖的,其酶-抑制剂解离半寿命分别>480分钟和270分钟。此外,GSK1070916还是一种与ATP竞争的抑制剂。[1]用GSK1070916处理的人类肿瘤细胞表现出剂量依赖性地抑制Histone H3第10丝氨酸的磷酸化,这是Aurora B的特异性底物。GSK1070916还能够抑制超过100种跨越广泛肿瘤类型细胞系的肿瘤细胞增殖,EC50值均<10 nM,中值EC50为8 nM。尽管GSK1070916对增殖细胞具有强大活性,但在原代、非分裂的正常人静脉内皮细胞中观察到其活性有显著变化。此外,GSK1070916处理的细胞并不在有丝分裂中停滞,而是失败分裂成为多倍体,最终导致凋亡。[2]在另一项研究中,还报告了高染色体数与对Aurora B和C抑制的抵抗性相关,这表明具有绕过高倍体检查点机制的细胞对GSK1070916具有抵抗性。[3] |
| 体内活性 | GSK1070916(25、50或100 mg/kg)在小鼠中显示出对Aurora B特异性底物磷酸化的剂量依赖性抑制作用。该化合物在包括乳腺癌、结肠癌、肺癌和两种白血病模型在内的10种人类肿瘤异种移植模型中展示了抗肿瘤效果。[2] |
| 存储条件 | Powder: -20°C for 3 years | In solvent: -80°C for 1 year | Shipping with blue ice/Shipping at ambient temperature. |
| 溶解度 | DMSO : 8.53 mg/mL (16.8 mM), Sonication is recommended.
Ethanol : 8 mg/mL (15.76 mM), Sonication is recommended.
10% DMSO+40% PEG300+5% Tween 80+45% Saline : 3.3 mg/mL (6.5 mM), Sonication is recommended.
H2O : < 1 mg/mL (insoluble or slightly soluble)
|
| 关键字 | Tie-2 | Tie2 | SIK | Inhibitor | inhibit | GSK-1070916 | GSK 1070916 | FLT1 | AuroraKinase | Aurora Kinase | Aurora C-INCENP | Aurora B-INCENP | Apoptosis |
| 相关产品 | Chitosan oligosaccharide | Urea | Dimethyl phthalate | Aceglutamide | Alginic acid | Cysteamine hydrochloride | Metronidazole | Dextran sulfate sodium salt (MW 5000) | Citric Acid Triammonium | Gum arabic | Stavudine | Tamoxifen |
| 相关库 | 抑制剂库 | 经典已知活性库 | 抗癌活性化合物库 | 已知活性化合物库 | 激酶抑制剂库 | 高选择性抑制剂库 | 抗衰老化合物库 | 膜蛋白靶向化合物库 | 药物功能重定位化合物库 | 酪氨酸激酶分子库 | 抗癌临床化合物库 | 抗癌药物库 |