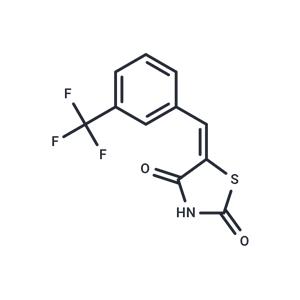
| 激酶实验 | Scintillation Proximity Assay: Methyltransferase activity assays are performed by monitoring the incorporation of tritiumlabeled methyl group from S-adenosylmethionine (3H-SAM) to biotinylated peptide substrates using Scintillation Proximity Assay (SPA) for PRC2-EZH2 trimeric complex (EZH2:EED:SUZ12), PRC2-EZH1 pentameric complex (EZH1:EED:SUZ12:RBBP4:AEBP2), SETD7, G9a, GLP, SETDB1, SETD8, SUV420H1, SUV420H2, SUV39H2, MLL1 tetrameric complex (MLL:WDR5:RbBP5:ASH2L), PRMT1, PRMT3, PRMT5-MEP50 complex and SMYD2. The reaction buffer for SMYD2 and SMYD3 is 50 mM Tris pH 9.0, 5 mM DTT, 0.01% TritonX-100; for G9a, GLP and SUV39H2 is 25 mM potassium phosphate pH 8.0, 1 mM EDTA, 2 mM MgCl2 and 0.01% Triton X-100; and for other HMTs 20 mM Tris pH 8.0, 5 mM DTT, 0.01% TritonX-100. To stop the enzymatic reactions, 10 μL of 7.5 M guanidine hydrochloride is added, followed by 180 μL of buffer, mixed and transferred to a 384-well FlashPlate. After mixing, the reaction mixtures are incubated and the CPM counts are measured using Topcount plate reader. The CPM counts in the absence of compound for each data set are defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set are defined as background (0%). IC50 values are determined using compound concentrations ranging from 100 nM to 100 μM. The IC50 values are determined using SigmaPlot software. EZH2-Y641F assays are performed using 30 nM of enzyme in 20 mM Tris pH 8, 5 mM DTT, 0.01% Triton X-100, 5 μM SAM and 1 μM of H3 (1-24) peptide (same as for the wild-type PRC2-EZH2 complex). For DNMT1, the assay is performed using hemimethylated dsDNA as a substrate. The dsDNA substrate is prepared by annealing two complementary strands (biotintlated forward strand: BGAGCCCGTAAGCCCGTTCAGGTCG and reverse strand: CGACCTGAACGGGCTTACGGGCTC), synthesized by Eurofins MWG Operon. Reaction buffer is 20 mM Tris-HCl, pH 8.0, 5 mM DTT, 0.01% Triton X-100.Methyltransferase activity assays for DOT1L is performed using Filter-plates. Reaction mixtures in 20 mM Tris-HCl, pH 8.0, 5 mM DTT, 2 mM MgCl2 and 0.01% Triton X-100 are incubated at room temperature for 1h, 100 μL 10% TCA is added, mixed and transferred to filter-plate. Plates are centrifuged at 2000 rpm for 2 min followed by 2 additional 10% TCA wash and one ethanol wash (180 μL) followed by centrifugation. Plates are dried and 100 μL MicroO is added and centrifuged. 70 μL MicroO is added and CPM are measured using Topcount plate reader. |

 United States
United States