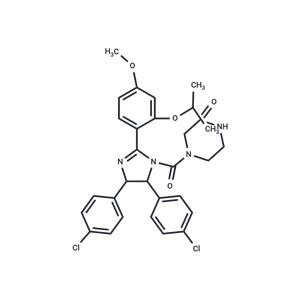
| Name | Nutlin-3 |
| Description | Nutlin-3 is an MDM2 antagonist that inhibits MDM2-p53 interaction (Ki=90 nM) and activates p53. Nutlin-3 binds preferentially to the p53-binding pocket of MDM2, leading to stabilization of p53 and activation of the p53 pathway. Nutlin-3 has antitumor activity. |
| Cell Research | Nutlin-3 (NUT) is dissolved with DMSO (100 mM) and diluted with appropriate media[2]. Human non-small-cell lung carcinoma wild type p53-containing H460 and A549, human non-small-cell lung carcinoma p53-null H1299, and human colon cancer HCT116 (p53+/+ and p53-/-) cells are used. Cells (1.5×105) are plated into 6-well plates, and incubated at 37°C overnight. After treatment of Inauhzin and Nutlin-3 at the indicated concentrations for 48 h, cells are harvested, fixed in 70% ice-cold ethanol overnight at -20°C, resuspended in propidium iodide-solution (50 μg/mL PI, 0.1 mg/mL RNase A, 0.05% Tritin X-100 in PBS) for 40 min at 37°C, then analyzed for DNA content using a flow cytometer and proprietary software[2]. |
| Kinase Assay | Biacore study: Competition assay is performed on a Biacore S51. A Series S Sensor chip CM5 is utilized for the immobilization of a PentaHis antibody for capture of the His-tagged p53. The level of capture is ~200 response units (1 response unit corresponds to 1 pg of protein per mm 2). The concentration of MDM2 protein is kept constant at 300 nM. Nutlin-3 is dissolved in DMSO at 10 mM and further diluted to make a concentration series of Nutlin-3 in each MDM2 test sample. The assay is run at 25°C in running buffer (10 mM Hepes, 0.15 M NaCl, 2% DMSO). MDM2-p53 binding in the presence of Nutlin-3 is calculated as a percentage of binding in the absence of Nutlin-3 and IC50 is calculated |
| In vitro | METHODS: 22RV1 (p53 WT), DU145 (p53 Mut) and PC-3 (p53 Null) cells were treated with Nutlin-3 (2-10 µM) for 48 h. Apoptosis was detected by Hoechst 33342 staining.
RESULTS: Nutlin-3 induced a significant increase in apoptosis in 22RV1 cells, whereas apoptosis was not significantly increased in p53-deficient DU145 or PC-3 cells. Nutlin-3 preferentially increased apoptosis in cells expressing WT p53. [1]
METHODS: H460 and HCT116 p53+/+ cells were treated with Nutlin-3 (0-100 µM) and Inauhzin (0-100 µM) for 72 h. Cell viability was measured by WST assay.
RESULTS: Inauhzin and Nutlin-3 showed significant synergistic effects in inhibiting the growth of both tumor cell lines, as the CI values were significantly less than 1 in the effective dose range, especially when lower doses of Nutlin-3 were used. [2] |
| In vivo | METHODS: To assay antitumor activity in vivo, Nutlin-3 (150 mg/kg, EtOH: Tween: 5% Glucose = 5:5:90; administered orally twice daily for four days and stopped for two days) and Inauhzin (15 mg/kg, intraperitoneally once daily) were administered to SCID mice bearing HCT116 p53+/+ xenografts for 21 days.
RESULTS: Each compound alone reduced mean tumor volume by less than 20% after 21 days of treatment. However, when both treatments were combined at the same dose, the final average tumor volume was reduced by 60%. inauhzin and Nutlin-3 activated p53 and inhibited the growth of xenograft tumors in the animals. [2] |
| Storage | Powder: -20°C for 3 years | In solvent: -80°C for 1 year | Shipping with blue ice. |
| Solubility Information | DMSO : 55 mg/mL (94.58 mM)
|
| Keywords | E3 ligating enzyme | E2 conjugating enzyme | E1/E2/E3 Enzyme | E1 activating enzyme | Ubiquitin conjugating enzyme | Nutlin-3 | inhibit | MDM-2/p53 | Nutlin 3 | Ubiquitin activating enzyme | Inhibitor | Ubiquitin ligase |
| Inhibitors Related | NSC697923 | WS-383 | PYZD-4409 | NAcM-OPT | RG7112 | PYR-41 | BC-1382 | NSC232003 | NAE-IN-M22 | Indole-3-carbinol | Idasanutlin | Navtemadlin |
| Related Compound Libraries | Anti-Pancreatic Cancer Compound Library | Bioactive Compound Library | Ubiquitination Compound Library | HIF-1 Signaling Pathway Compound Library | Inhibitor Library | Stem Cell Differentiation Compound Library | Anti-Aging Compound Library | Bioactive Compounds Library Max | Human Metabolite Library | Anti-Cancer Active Compound Library |

 United States
United States